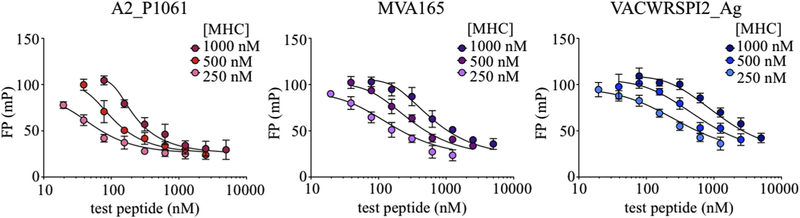
Fig. 7. Fitting competitive binding inhibition curves using a cubic equation.
To determine Ki values for the A2_P1061, MVA165, and VACWRSPI2_Ag peptides, three curves obtained at different [MHC] were simultaneously fit using an implicit cubic equation (Appendix C). Best fit Ki values obtained with Kd = 16 nM and fract = 0.2 shown in Table 3, along with a range of Ki values obtained with reasonable parameter values (mPfree = 22 to 28, mPbound = 115 to 150, Kd = 10–20 nM, and fract = 0.08 to 0.2).