Abstract
In plants, nutrient transporters require tight regulation to ensure optimal uptake in complex environments. The activities of many nutrient transporters are post-translationally regulated by reversible phosphorylation, allowing rapid adaptation to variable environmental conditions. Here, we show that the Arabidopsis root epidermis-expressed ammonium transporter AtAMT1;3 was dynamically (de-)phosphorylated at multiple sites in the cytosolic C-terminal region (CTR) responding to ammonium and nitrate signals. Under ammonium resupply rapid phosphorylation of a Thr residue (T464) in the conserved part of the CTR (CTRC) effectively inhibited AtAMT1;3-dependent NH4+ uptake. Moreover, phosphorylation of Thr (T494), one of three phosphorylation sites in the non-conserved part of the CTR (CRTNC), moderately decreased the NH4+ transport activity of AtAMT1;3, as deduced from functional analysis of phospho-mimic mutants in yeast, oocytes, and transgenic Arabidopsis. Double phospho-mutants indicated a role of T494 in fine-tuning the NH4+ transport activity when T464 was non-phosphorylated. Transient dephosphorylation of T494 with nitrate resupply closely paralleled a transient increase in ammonium uptake. These results suggest that T464 phosphorylation at the CTRC acts as a prime switch to prevent excess ammonium influx, while T494 phosphorylation at the CTRNC fine tunes ammonium uptake in response to nitrate. This provides a sophisticated regulatory mechanism for plant ammonium transporters to achieve optimal ammonium uptake in response to various nitrogen forms.
Keywords: Ammonium transporter (AMT), ammonium uptake, membrane transport, multisite phosphorylation, nitrogen signals, phosphorylation, post-translational regulation
Phosphorylation/dephosphorylation at multiple sites in the C-terminal region of AtAMT1;3 regulates ammonium transporter activity, illustrating a novel regulatory mechanism to control root ammonium uptake in response to different nitrogen signals.
Introduction
Ammonium is a major nitrogen (N) form in most soils and is preferentially taken up, providing an essential N metabolite in plants (Marschner and Rengel, 2012). Ammonium may also act as a nutrient signal molecule to trigger physiological and morphological responses in plants (Lima et al., 2010; Liu and von Wirén, 2017). High-affinity ammonium uptake and root-to-shoot ammonium translocation are mainly mediated by ammonium transporters (AMTs), which belong to the AMT/MEP/Rh protein superfamily (Loqué and von Wirén, 2004). In Arabidopsis roots, the epidermis-expressed AtAMT1;1 and AtAMT1;3 and the endodermis-expressed AtAMT1;2 control uptake of ammonium via the symplastic and apoplastic transport pathway, respectively (Kaiser et al., 2002; Loqué et al., 2006; Mayer et al., 2006; Mayer and Ludewig 2006; Neuhäuser et al., 2007; Yuan et al., 2007a; Duan et al., 2018). AtAMT2;1 shows marginal expression in the epidermis, but is preferentially localized to the pericycle cells under elevated ammonium supply and is involved in the root-to-shoot translocation of ammonium (Neuhäuser et al., 2009; Giehl et al., 2017).
To respond to external N availability and the internal plant N status, AMT gene expression is tightly controlled in roots at multiple regulatory levels. At the transcriptional level, ammonium supply can either up- or down-regulate expression of AMTs, which is probably triggered by a downstream metabolite of ammonium, i.e. glutamine (Gazzarrini et al., 1999; Rawat et al., 1999; von Wirén et al., 2000; Sonoda et al., 2003a,b; Gu et al., 2013; Wu et al., 2015). The transcription factors of nitrate-inducible GARP-type transcriptional repressor 1 (NIGT1)/hypersensitive to low Pi-elicited primary root shortening 1 (HRS1) negatively regulate expression of AtAMT1;1, while the rice transcription factors growth regulation factor 4 (GRF4) and indeterminate domain 10 (IDD10) activate expression of OsAMT1;1 and OsAMT1;2, respectively (Xuan et al., 2013; Kiba et al., 2018; Li et al., 2018). At the post-transcriptional level, AtAMT1;1 mRNA turnover is strictly controlled by plant N availability (Yuan et al., 2007b).
To cope with elevated external ammonium, Arabidopsis AMT1-type ammonium transporters (AtAMT1s) can be post-translationally modified via phosphorylation of their cytosolic C-terminal region (CTR) to shut off their transport activity (Loqué et al., 2007; Neuhäuser et al., 2007; Lanquar et al., 2009; Yuan et al., 2013). Within the homo- or hetero-trimeric complexes of AMT1s, the CTR interacts with a cytosolic loop of its own or the adjacent monomer (Khademi et al., 2004; van den Berg et al., 2016). The phosphorylation of a conserved Thr residue in the CTR (ThrCTR) can lead to trans-inactivation of the whole complex in an allosteric manner (Loqué et al., 2007; Neuhäuser et al., 2007; Yuan et al., 2013). The ThrCTR of AtAMT1;1 (T460) and AtAMT1;2 (T472) are phosphorylated by the calcineurin B-like protein-interacting protein kinase 23 (CIPK23)–calcineurin B-like protein 1 (CBL1) complex, while OsAMT1;2 (T453) is phosphorylated by the serine/threonine/tyrosine (STY) kinase ACT domain protein kinase 1 (ACTPK1) (Straub et al., 2017; Beier et al., 2018). In both cases, phosphorylation is triggered by an ammonium signal. Phosphorylation of ThrCTR in AtAMT1;3 (T464) has also been detected in vivo by phosphoproteomics and is able to regulate the transporter activity (Yuan et al., 2013; Menz et al., 2016). However, AtAMT1;3 T464 is not the target of AtCIPK23–CBL1, even though its conserved phosphorylation target site is indistinguishable from those of other AtAMT1s by sequence (Straub et al., 2017). Besides ThrCTR, other phosphorylation sites in the CTRs of AtAMT1;1 and AtAMT1;3 have been identified by previous phosphoproteomic studies in vivo (Lanquar et al., 2009; Engelsberger and Schulze, 2012; Menz et al., 2016). Notably, distinct dynamic phosphorylation patterns on these multiple sites were observed in plants in response to changes in the N nutritional status, including supplies of different forms of N. Whether these multiple phosphorylation sites are functionally involved in regulating AMT1s transporter activity and how they differentially respond to variable N supply, however, remain to be elucidated.
Here, we assessed the correlations between 15N-labeled high-affinity ammonium influx and multisite phosphorylation status of AtAMT1;3 CTR in Arabidopsis roots, and showed that ammonium resupply-triggered phosphorylation of T464 in the conserved part of the CTR rapidly decreased ammonium uptake. However, nitrate resupply led to a transient dephosphorylation of T494 in the non-conserved part of the CTR, accompanied by an increase in ammonium uptake. Using yeast, oocytes, and transgenic Arabidopsis, AtAMT1;3 T464, T494 single or double phospho-mimic variants generated by site-directed mutagenesis were functionally analysed in vitro and in vivo. The results indicated that T494 phosphorylation could partially decrease AtAMT1;3 transporter activity and this fine-tuning role of T494 relied on the non-phosphorylation status of T464. These findings thus extend our understanding of AtAMT1s phospho-dependent regulation from single to multiple site levels, and highlight the mechanisms by which these multisite phosphorylation/dephosphorylation processes cooperate to achieve optimal uptake of ammonium under control of different N signals.
Materials and methods
Vector constructs
The open reading frame of AtAMT1;3 (AtAMT1;3-ORF) was cloned into vector pTOPO (Invitrogen) and sub-cloned into yeast expression vector p426-HXT7 through the EcoRI site, yielding the plasmid p426-AtAMT1;3. Mutations were introduced into AtAMT1;3-ORF by using the Q5® Site-Directed Mutagenesis Kit (NEB) with specific primers (Supplementary Table S1 at JXB online) and verified by sequencing, yielding the plasmids p426-AtAMT1;3 T464A, p426-AtAMT1;3 T464D, p426-AtAMT1;3 H474stop, p426-AtAMT1;3 S480A, p426-AtAMT1;3 S480D, p426-AtAMT1;3 S487A, p426-AtAMT1;3 S487D, p426-AtAMT1;3 T494A, p426-AtAMT1;3 T494D, p426-AtAMT1;3 T464A T494A, p426-AtAMT1;3 T464A T494D, p426-AtAMT1;3 T464D T494A, p426-AtAMT1;3 T464D T494D, p426-AtAMT1;3 T464A S480A S487A, and p426-AtAMT1;3 T464A S480D S487D for yeast transformation.
The AtAMT1;3 promoter (AtAMT1;3pro) was amplified by PCR from the pBI101-AtAMT1;3pro::AtAMT1;3::GFP construct (Loqué et al., 2006) using the specific primers AtAMT1;3pro-XbaI and AtAMT1;3pro-ApaI (Supplementary Table S1). The amplified fragment was digested by XbaI and ApaI, and cloned into vector pT-Hygromycin to generate the pT-AtAMT1;3pro construct. The AtAMT1;3-ORF and AtAMT1;3 phospho-mutants were amplified by PCR using specific primers (Supplementary Table S1) and cloned into the pT-AtAMT1;3pro construct through the ApaI site, resulting in the plasmids pT-AtAMT1;3pro::AtAMT1;3, pT-AtAMT1;3pro::AtAMT1;3 T464A, pT-AtAMT1;3pro::AtAMT1;3 T494A, pT-AtAMT1;3pro::AtAMT1;3 T494D, pT-AtAMT1;3pro::AtAMT1;3 T464A T494A, and pT-AtAMT1;3pro::AtAMT1;3 T464A T494D for Arabidopsis transformation.
The AtAMT1;3-ORF and AtAMT1;3 phospho-mutants were amplified by PCR with specific primers (Supplementary Table S1) and cloned into the oocyte expression vector pOO2 through SpeI and EcoRI sites (Baukrowitz et al., 1999), yielding the plasmids pOO2-AtAMT1;3, pOO2-AtAMT1;3 T494A, pOO2-AtAMT1;3 T494D, pOO2-AtAMT1;3 T464A, pOO2-AtAMT1;3 T464A T494A, and pOO2-AtAMT1;3 T464A T494D for oocyte assays.
Growth complementation and 15N-labeled ammonium uptake assays in yeast
The plasmids were heated shock-transformed in triple-Δmep (1–3) deletion yeast strain 31019b (Marini et al. 1997), and the transformants were selected on solid YNB medium with arginine (Arg) (2% Agar, YNB without amino acids and ammonium sulfate (Difco), 3% glucose and 0.1% Arg). A growth complementation assay was performed on solid YNB medium supplemented with 3% glucose, 1 mM NH4Cl or 1 mM Arg as the sole nitrogen source, buffered with 50 mM MES/Tris (pH 5.5). The 15N-labeled ammonium uptake assay was performed at a concentration of 250 μM 15NH4+ (99.12 atom% 15N) as described previously by Loqué et al. (2009).
Electrophysiological measurements, preparation and injection of oocytes
The electrophysiological methods used were described by Mayer and Ludewig (2006). The oocytes were from Ecocyte Bioscience (Castrop-Rauxel), presorted again and injected with 50 nl of cRNA (1 µg µl−1). Oocytes were kept in ND96 for 4 d at 18 °C and then placed in a small recording chamber. The recording solution was 110 mM choline chloride, 2 mM CaCl2, 2 mM MgCl2, and 5 mM MES, pH adjusted to 5.5 with Tris. Variable ammonium concentrations were added as NH4Cl salt. Currents without added ammonium were subtracted at each voltage.
Generation of transgenic Arabidopsis plants
Arabidopsis mutant qko (atamt1;1, atamt1;2, atamt1;3 and atamt2;1) was used for transformation (Yuan et al., 2007a). Agrobacterium-mediated transformation was performed using Agrobacterium tumefaciens strain GV3101 by the standard floral-dip method (Clough and Bent, 1998). Transgenic lines were selected on half-strength Murashige & Skoog medium containing 25 mg l−1 hygromycin B. Independent T3 homozygotes with a single-copy transgene were used for the analysis. Plants were grown in a climate-controlled greenhouse under 16/8 h light/dark cycle at temperature 22 °C/18 °C.
15N-labeled ammonium influx assay in roots
Arabidopsis plants were cultured hydroponically as described by Loqué et al. (2006). Six-week-old Arabidopsis plants were grown hydroponically in a growth cabinet at 22 °C with 10 h light/14 h dark photoperiod illuminated at 100 μmol m−2 s−1. After 4 d N starvation, 4 mM NH4Cl or 4 mM KNO3 was used for ammonium or nitrate resupply treatment, respectively. All nutrient solutions were adjusted to pH 5.8. Nutrient solutions were renewed every other day. Influx of 15N-labeled NH4+ into roots of Arabidopsis plants was measured after rinsing the roots of hydroponically grown plants in 1 mM CaSO4 solution for 1 min, followed by an incubation for 6 min in full nutrient solution (pH 5.8) containing 250 μM concentrations of 15N-labeled NH4+ (99.12 atom% 15N) as the sole N source, and a final wash in 1 mM CaSO4 solution for 1 min. Then roots were harvested and freeze-dried at −50 °C. A 1 mg aliquot of ground samples was used for 15N determination by isotope mass spectrometry (DELTAplus XP, Thermo-Finnigan).
RNA extraction and quantitative RT-PCR analysis
Total RNA was extracted from roots of hydroponic grown plants using RNAiso Plus (Takara). RNA samples were pretreated with gDNA Eraser of the PrimeScript™ RT reagent Kit (Takara) and reverse transcribed into cDNA. Using the IQ5 Real-Time PCR System (Bio-Rad), quantitative RT-PCR (qPCR) was performed with SYBR Green. Expression of AtUBQ10 and AtEIF4a (Boudsocq et al., 2010) was used for normalizing gene expression. The specific primers used in the qPCR analysis are listed in Supplementary Table S1.
Protein gel blot analysis
The microsomal membrane fraction was isolated from Arabidopsis roots as described by Yuan et al. (2007a). AtAMT1;3 polyclonal antibodies were produced against peptide sequences at the C-terminus (n-DPGSPFPRSATPPRV-c) (Beijing Protein Innovation Co., Ltd); the AtAMT1s TCTR phosphorylation specific antibody (AtAMT1s TCTR-P) was descried by Straub et al. (2017). Protein (15 μg) was denatured with loading buffer at 37 °C for 30 min, separated on 10% SDS polyacrylamide gels, and transferred to a polyvinylidene difluoride (PVDF) membrane (Amersham Hybond-P; 0.45 μm; GE Healthcare) by electroblotting. Protein was stained with Coomassie Brilliant Blue as the loading control. The PVDF membrane was blocked by incubation in PBST with 5% skim milk powder for 1 h at room temperature and incubated with primary antibodies (1:5000 for anti-AtAMT1;3 and 1:500 for anti-AtAMT1s TCTR-P) and secondary antibodies (1:10 000) followed by ECL detection (Amersham ECLTM Advance Western Blotting Detection Kit). MagicMarkTM XP Western Standard (Invitrogen) and Blue Plus IV Protein Marker (TransGen Biotech) were used to indicate the molecular masses of proteins.
Results
AtAMT1;3-dependent ammonium uptake in Arabidopsis roots was rapidly repressed by resupply of ammonium, but transiently induced by nitrate
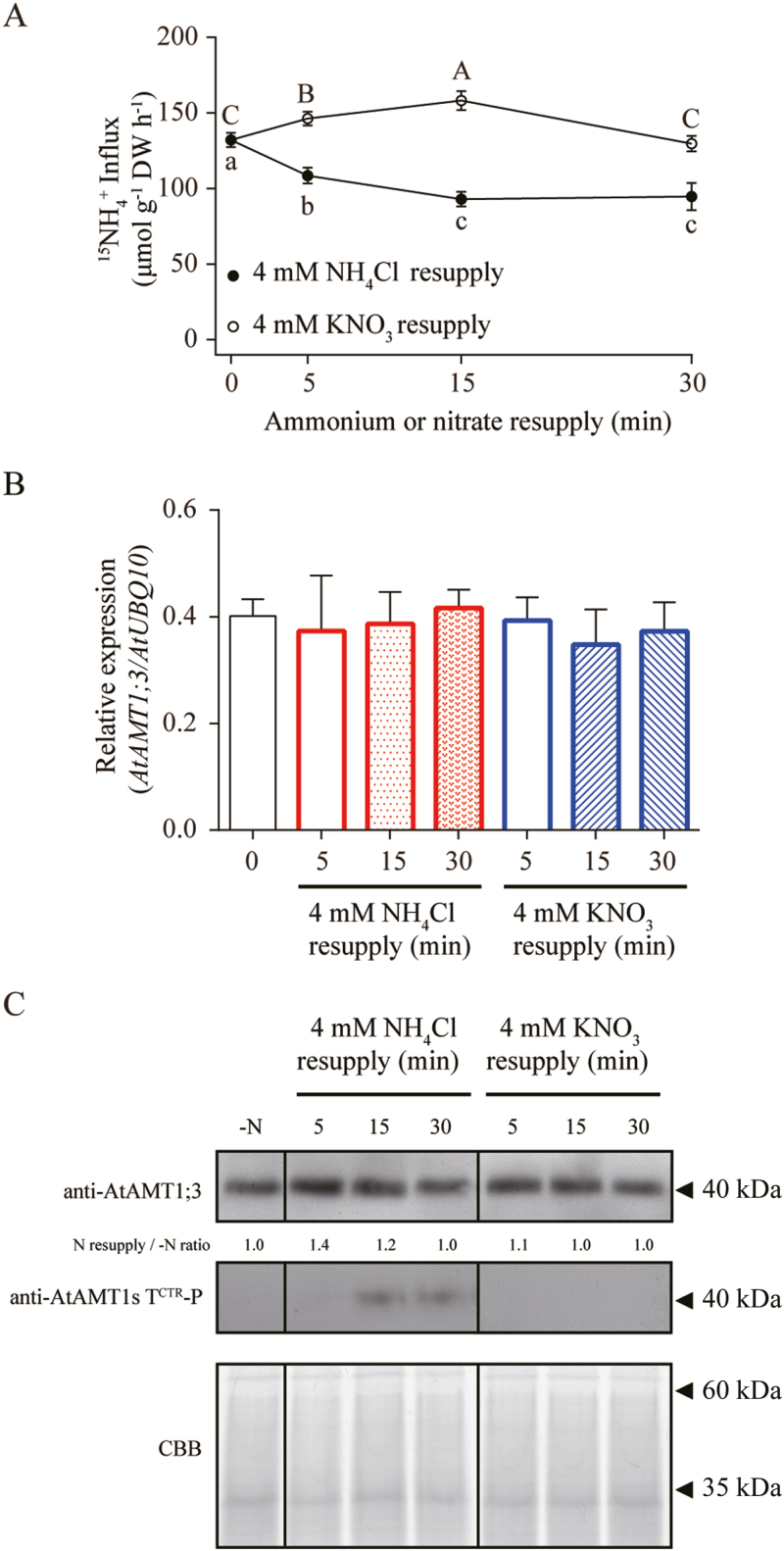
To investigate whether AtAMT1;3 transporter activity in roots differentially responds to resupply of ammonium or nitrate, we employed the Arabidopsis mutant qko+13 in which AtAMT1;3 expression was reconstituted in the amt-qko background (atamt1;1, atamt1;2, atamt1;3, atamt2;1), allowing us to quantify AtAMT1;3-dependent ammonium uptake capacity due to the disruption of three other endogenous AtAMT genes (Yuan et al., 2007a). Hydroponically grown qko+13 seedlings were starved of N, and resupplied with 4 mM ammonium or nitrate for different time periods. With these plants, a short-term high-affinity 15N-labeled ammonium influx assay was performed in roots. AtAMT1;3 expression was simultaneously quantified at the transcript level by qPCR and at the protein level by western blot analysis (Fig. 1). Ammonium influx rates in roots of qko+13 were rapidly decreased up to approx. 30% within 15 min of ammonium resupply compared with that under N deficiency (Fig. 1A). With resupply of nitrate, however, ammonium influx rates were transiently induced up to 17% within 15 min and then returned to the original level after 30 min. Compared with that in N-deficient roots, the abundance of AtAMT1;3 transcripts did not show any change after resupply of ammonium or nitrate (Fig. 1B; Supplementary Fig. S1). Similarly, no change was observed at the protein levels as revealed by western blot analysis using an AtAMT1;3-specific antibody (Fig. 1C). Thus, these results revealed distinct dynamic patterns for AtAMT1;3 activity changes in response to different N forms. This regulation was at the post-translational level, because the transcript and protein abundances of AtAMT1;3 did not change accordingly.
Fig. 1.
Dynamic patterns of AtAMT1;3-dependent root ammonium uptake and T464 phosphorylation in response to ammonium and nitrate resupply. Six-week-old hydroponically grown Arabidopsis mutant qko+13 (atamt1;1, atamt1;2 and atamt2;1) were subjected to N starvation for 4 d, and then resupplied with 4 mM NH4Cl or 4 mM KNO3 for 5, 15, and 30 min. (A) 15N-labeled ammonium influx in roots determined at the concentration of 250 μM. (B) Transcript expression levels of AtAMT1;3 in roots quantified by qPCR with three replicates, and normalized by AtUBQ10 expression level. (C) Protein expression levels of AtAMT1;3 and phosphorylated AtAMT1;3 at T464 in roots determined by western blot analysis using anti-AtAMT1;3 and anti-AMT1s ThrCTR-P antibody, respectively, normalized by Coomassie Brilliant Blue staining. Bars indicate means ±SD (n=3) and significant differences at P<0.001 according to Tukey’s test are indicated by different letters. (This figure is available in color at JXB online.)
Ammonium signals triggered AtAMT1;3 phosphorylation at T464 in the CTRC, whereas nitrate signals triggered multisite (de-)phosphorylation in the CTRNC
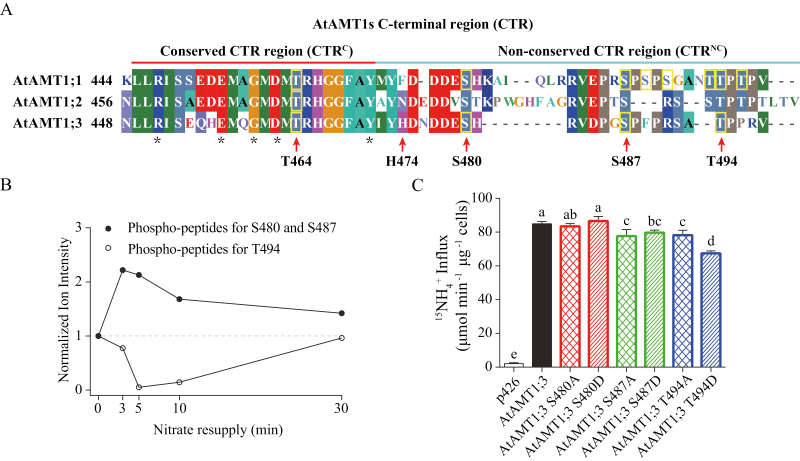
Alignment of AtAMT1s amino acids sequences showed that their CTR consisted of a conserved (CTRC) and a non-conserved (CTRNC) part, which is a common feature for AMT/MEP family members (Fig. 2A; Loqué et al., 2007). By collecting the phosphorylated peptide sequences available from previous phosphoproteomics or phospho-specific antibody analyses of different Arabidopsis organs under various treatments, the number of in vivo phosphorylated Ser/Thr residues identified in the CTR was eight for AtAMT1;1, one for AtAMT1;2, and four for AtAMT1;3 (Supplementary Table S2). In the CTRC, only a single conserved phosphorylated residue (ThrCTR) appeared (AtAMT1;1 T460/AtAMT1;2 T472/AtAMT1;3 T464) (Fig. 2A). In the CTRNC, however, multiple phosphorylation sites were present in AtAMT1;1 (seven residues) and AtAMT1;3 (three residues), particularly two conserved Ser (AtAMT1;1 S475/AtAMT1;3 S480, AtAMT1;1 S488/AtAMT1;3 S487) and one conserved Thr (AtAMT1;1 T497/AtAMT1;3 T494). Apart from the well-characterized ThrCTR, multiple phosphorylation sites resided in the CTRNC, but potential functional roles in regulating ammonium transporters are unknown.
Fig. 2.
Phosphorylation dynamics of AtAMT1;3 CTRNC at multiple sites (S480, S487, and T494) in response to nitrate and functional analysis of corresponding phospho-mutants in yeast. (A) In vivo phosphorylation sites in the CTR of AtAMT1s identified by phosphoproteomics and phospho-specific antibodies (Supplementary Table S1). Alignment of amino acid sequences showed that the CTR of AtAMT1s consisted of a conserved (CTRC) and a non-conserved (CTRNC) part that are underlined in red and blue, respectively. The phosphorylated Thr/Ser residues are labeled with a yellow box. Five universally conserved residues within the CTRC, including the ‘ExxGxD’ motif, are labeled with asterisks. (B) Normalized ion intensity of phosphorylated AtAMT1;3 peptides HGGFAYIYHDNDDES(ph)HRVDPGS(ph)PFPR (S480/S487) and SAT(ph)PPRV (T494) that responded to nitrate. The data were obtained from Engelsberger and Schulze (2012). (C) Influx of 15N-labeled ammonium into yeast mutant 31019b expressing empty vector p426, AtAMT1;3 wild type, and different AtAMT1;3 phospho-variants: S480A, S480D, S487A, S487D, T494A, and T494D. Influx assays were performed at the concentration of 250 μM. Bars indicate means ±SD (n=5) and significant differences at P<0.001 according to Tukey’s test are indicated by different letters.
To test whether resupply of ammonium can trigger AtAMT1;3 T464 phosphorylation in the CRTC and thereby inhibit ammonium uptake capacity in qko+13 roots, a phospho-specific antibody against ThrCTR of AtAMT1s (Straub et al., 2017) was employed for western blot analysis (Fig. 1C). Due to an exclusive expression of AtAMT1;3 in qko+13 roots, the band detected by this antibody reflects phosphorylation of AtAMT1;3 at T464. Compared with N-deficient roots, the phosphorylated polypeptides appeared after 15 min resupply of ammonium (Fig. 1C) and coincided with decreases of AtAMT1;3-dependent ammonium uptake capacity (Fig. 1A). No T464 phosphorylation, however, was detected under nitrate resupply. Because T464 phosphorylation inactivates AtAMT1;3 transporter activity (Yuan et al., 2013), this result suggested that ammonium signals, but not nitrate, selectively induced T464 phosphorylation and thereby inhibited root ammonium uptake capacity.
With resupply of nitrate, but not ammonium, considerable phosphorylated Ser/Thr residues in the CTRNC of AtAMT1;3 were identified by previous phosphoproteomics of Arabidopsis plants (Engelsberger and Schulze, 2012; Fig. 2B; Supplementary Fig. S2). Among them, two Ser residues (S480/S487) were simultaneously phosphorylated, and the phosphorylation levels were increased up to 2-fold after 3 min resupply of nitrate to N-deficient seedlings. The phosphorylated Thr residue (T494), by contrast, was rapidly dephosphorylated within 5 min under nitrate resupply, and then returned to the original level after 30 min. Thus, a close correlation between dynamic changes of S480/S487/T494 phosphorylation status and ammonium uptake rates in roots of qko+13 (Fig. 1A) may suggest that nitrate signals could control multisite phosphorylation/dephosphorylation in the CTRNC for regulating AtAMT1;3 transporter activity.
T494 phosphorylation in the CTRNC was involved in regulating AtAMT1;3 transporter activity in vitro and in vivo
To verify whether phosphorylation sites in the CTRNC are involved in regulating AtAMT1;3 transporter activity, mutants mimicking non-phosphorylation (Ser/Thr replaced by Ala) and phosphorylation (Ser/Thr replaced by Asp) of each site were generated (Fig. 2C). These AtAMT1;3 phospho-variants together with the wild type were then functionally analysed in the yeast strain 31019b (triple-mepΔ), which grows poorly on <5 mM ammonium as a sole N source (Marini et al., 1997). The yeast cells expressing all variants were able to confer growth complementation on 1 mM ammonium, and no visible growth difference was observed between the wild type and phospho-mutants (Supplementary Fig. S3). By quantitative analysis using a short-term 15NH4+ uptake assay, it was found that the mutations at S487 and T494, but not S480, resulted in significant decreases of transporter activity compared with that of the wild type (Fig. 2C). The S487 phospho-mutants showed approx. 8% reduction, but no difference was observed between the non-phosphorylated (S487A) and phosphorylated form (S487D). However, compared with the non-phosphorylation mimic T494A, the phosphorylated mimic T494D revealed a significantly lower transporter activity (~10%), suggesting that AtAMT1;3 transporter activity is modulated via the T494 phosphorylation/dephosphorylation modification. Under nitrate resupply, the T494 dephosphorylation (derepressed activity) occurred within 5 min, closely paralleling the enhanced ammonium uptake rates in roots of qko+13 (Fig. 1A). This was followed by T494 phosphorylation (repressed activity) after 30 min, when the uptake had returned to the original level. Among these multisite phosphorylations, T494 phospho-modification was controlled by nitrate signals for regulating AtAMT1;3-dependent ammonium uptake in roots.
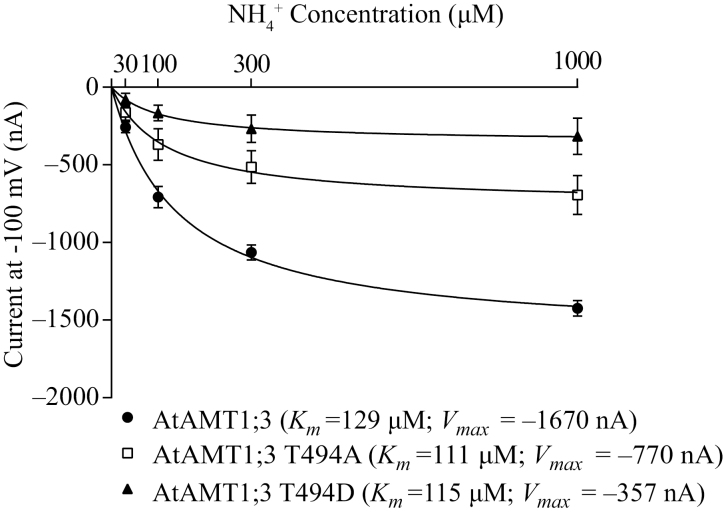
The AtAMT1;3 T494 phospho-mutants together with the wild type were then functionally characterized in Xenopus oocytes (Fig. 3). At −100 mV, oocytes expressing AtAMT1;3 T494A and T494D variants showed NH4+-specific inward currents resulting from electrogenic transport of ammonium (NH4+) into the oocytes. These ammonium-induced currents saturated in response to external ammonium concentrations that ranged from 30 to 1000 μM. The concentration needed to achieve half-maximal current (Km) was similar among AtAMT1;3 (129 μM), T494A (111 μM) and T494D (115 μM). However, compared with the maximal mean current (Vmax) of the AtAMT1;3 wild type (−1670 nA), the T494A and T494D mutants had a considerably lower Vmax of −770 and −357 nA, respectively. Transporter activity of the phosphorylation-mimic variant (T494D) was half that of the non-phosphorylation mimic (T494A). This result suggested that T494 phosphorylation was able to down-regulate AtAMT1;3 transporter activity without affecting the substrate affinity.
Fig. 3.
Functional characterization of AtAMT1;3 T494 phospho-mutants in Xenopus oocytes. Kinetic properties of ammonium-induced currents in oocytes injected with AtAMT1;3, and AtAMT1;3 phospho-mutants T494A and T494D. Inward currents were plotted under conditions of 30~1000 μM NH4Cl at −100 mV. Bars indicate means ±SD (n≥4). From a fit with the Michaelis–Menten equation, Vmax and Km were calculated.
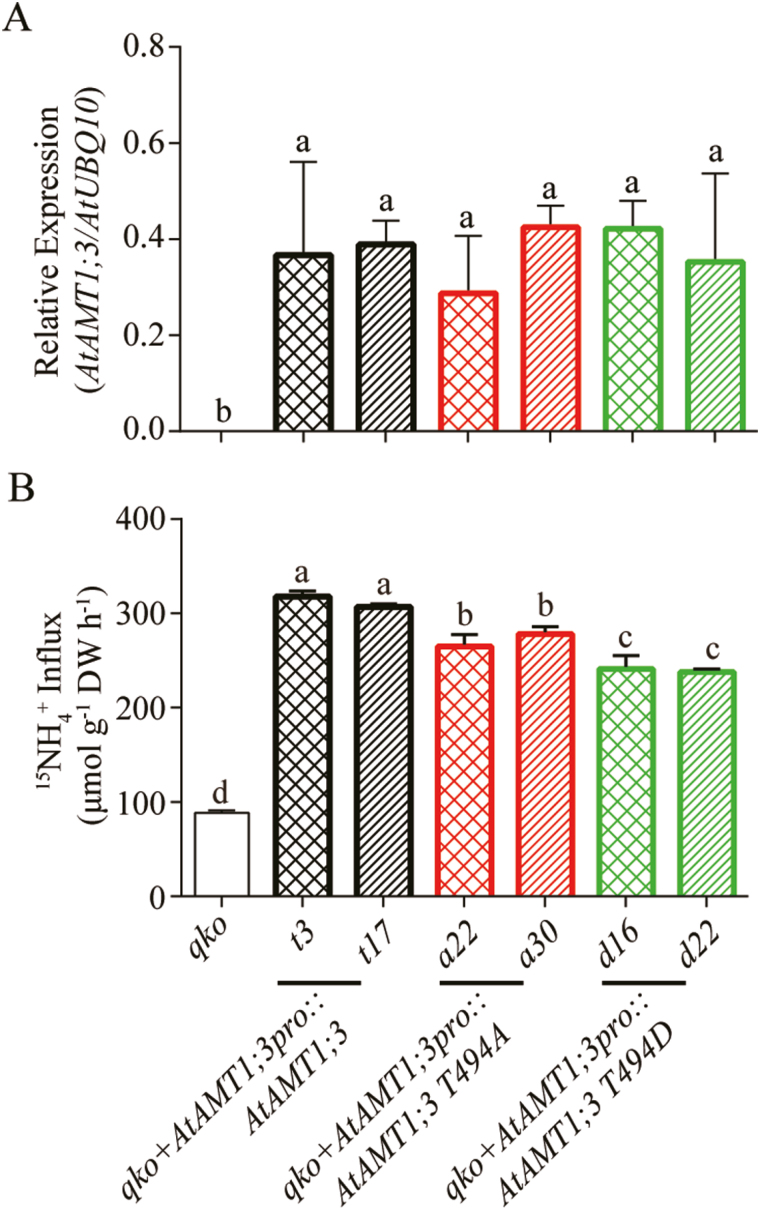
To further confirm whether T494 phosphorylation can regulate AtAMT1;3-dependent ammonium uptake in vivo, the AtAMT1;3 T494 phospho-variants together with the wild type were expressed in the Arabidopsis qko mutant background under the control of AtAMT1;3 native promoter (AtAMT1;3pro) (Fig. 4). Two independent homozygous lines of each construct, qko+AtAMT1;3pro::AtAMT1;3 (t3 and t17), qko+AtAMT1;3pro::AtAMT1;3 T494A (a22 and a30), and qko+AtAMT1;3pro::AtAMT1;3 T494D (d16 and d22), were selected, which had similar expression levels to transgenic AtAMT1;3 variants (Fig. 4A). The short-term 15N-labeled ammonium influx assay was then performed in N-deficient roots at a concentration of 250 μM, allowing the quantitative evaluation of transport capacity of AtAMT1;3 variants in vivo (Fig. 4B). The qko is defective in four endogenous AtAMT genes (AtAMT1;1, AtAMT 1;2, AtAMT 1;3, and AtAMT 2;1) and loses up to 90% of high-affinity ammonium uptake capacity in roots (Yuan et al., 2007a). Compared with qko, the transgenic lines expressing AtAMT1;3 T494A or T494D showed significantly enhanced ammonium influx rates. The in vivo transport activity of each variant was further determined after subtraction of the residual background transport activity of qko. Compared with the AtAMT1;3 wild type, T494A and T494D showed reduced activities of up to 18 and 33%, respectively (Fig. 4B). Similar to yeast and oocytes, the T494 phosphorylated form (T494D) of AtAMT1;3 conferred an 18% lower ammonium uptake rate than that of the non-phosphorylation mimic T494A. These results suggest that in vivo phospho-dependent modification of AtAMT1;3 at T494 could moderately regulate ammonium influx rates in roots.
Fig. 4.
Functional characterization of AtAMT1;3 T494 phospho-mutants in transgenic Arabidopsis roots. (A) Transcript expression levels of AtAMT1;3 in roots of qko, qko+AtAMT1;3pro::AtAMT1;3, qko+AtAMT1;3pro::AtAMT1;3 T494A or qko+AtAMT1;3pro::AtAMT1;3 T494D transgenic lines. Levels were quantified by qPCR with three replicates and normalized by AtUBQ10 expression. (B) Influx of 15N-labeled ammonium into roots of the same line as in (A). N-deficient roots were employed for 15N-labeled ammonium influx assays as determined at the concentration of 250 μM. Bars indicate means ±SD (n≥5) and significant differences at P<0.001 according to Tukey’s test are indicated by different letters. (This figure is available in color at JXB online.)
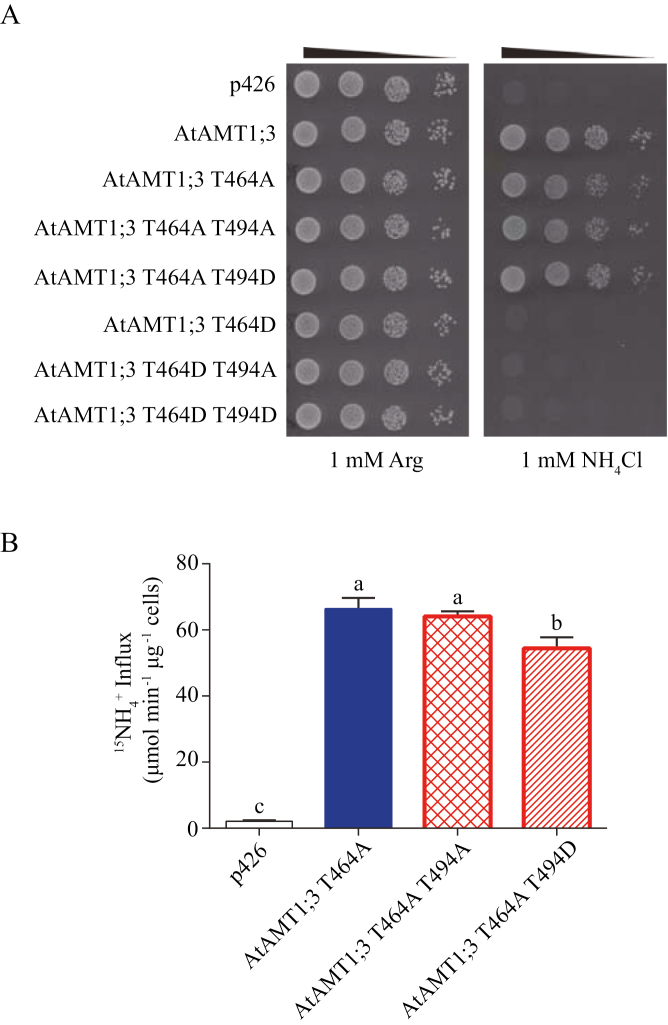
Non-phosphorylation of T464 in the CTRC was a prerequisite of fine-tuning AtAMT1;3 transport activity via T494 (de-)phosphorylation in the CTRNC
Given that phosphorylation of T464 in the CTRC and T494 in the CTRNC could negatively regulate the transport activity of AtAMT1;3 to varying extents, the possible interaction between these two phospho-dependent modifications was investigated using the corresponding double phospho-mutants (Fig. 5). As in a previous study (Yuan et al., 2013), yeast cells expressing the non-phosphorylation mimic variant T464A, but not the phosphorylation mimic variant T464D, were able to mediate growth under 1 mM ammonium (Fig. 5A). In the T464D background, neither phosphorylation mimic (T464D T494D) nor non-phosphorylation mimic (T464D T494A) double mutants conferred growth, resembling the single mutant T464D. In the T464A background, although all double mutants conferred yeast growth under low ammonium, the 15NH4+ uptake assay showed that the T464A T494A double mutant had a similar activity to the single mutant T464A, while the T464A T494D double mutant revealed approx. 27% reduction of transporter activity (Fig. 5B). When expressed in oocytes, the single non-phospho-mutant T464A of AtAMT1;3 revealed unexpectedly low activity (Supplementary Fig. S4), similar to the non-phospho-mutant of AtAMT1;2 at the equivalent site (T472A) (Neuhäuser et al., 2007). Nevertheless, by adding non-phosphorylation mimic T494 to the T464A mutant in the double phospho-variant T464A T494A, the transporter activity was restored. This indicated that, in the presence of non-phosphorylated T464, T494 dephosphorylation enhanced AtAMT1;3 transporter activity. Dephosphorylation/phosphorylation modifications of AtAMT1;3 at T464 apparently turned on/off transport activity, while those modifications at T494 moderately fine-tuned transporter activity as long as T464 was not phosphorylated.
Fig. 5.
Functional characterization of AtAMT1;3 T464 T494 double phospho-mutants in yeast. (A) Growth complementation of the yeast mutant 31019b (Δmep1-3) expressing empty vector p426, AtAMT1:3 wild type, AtAMT1;3 single phospho-variant T464A or T464D, double phospho-variants T464A T494A, T464A T494D, T464D T494A, or T464D T494D. Yeast cells were spotted on YNB medium supplemented with either 1 mM Arg or 1 mM ammonium as the sole N source at pH5.5 for 3 d at 28 °C. (B) Influx of 15N-labeled ammonium into yeast mutant 31019b expressing empty vector p426, AtAMT1;3 wild type, AtAMT1;3 single phospho-variant T464A and double phospho-variants T464A T494A or T464A T494D. Influx assays were performed at the concentration of 250 μM. Bars indicate means ±SD (n=5) and significant differences at P<0.001 according to Tukey’s test are indicated by different letters. (This figure is available in color at JXB online.)
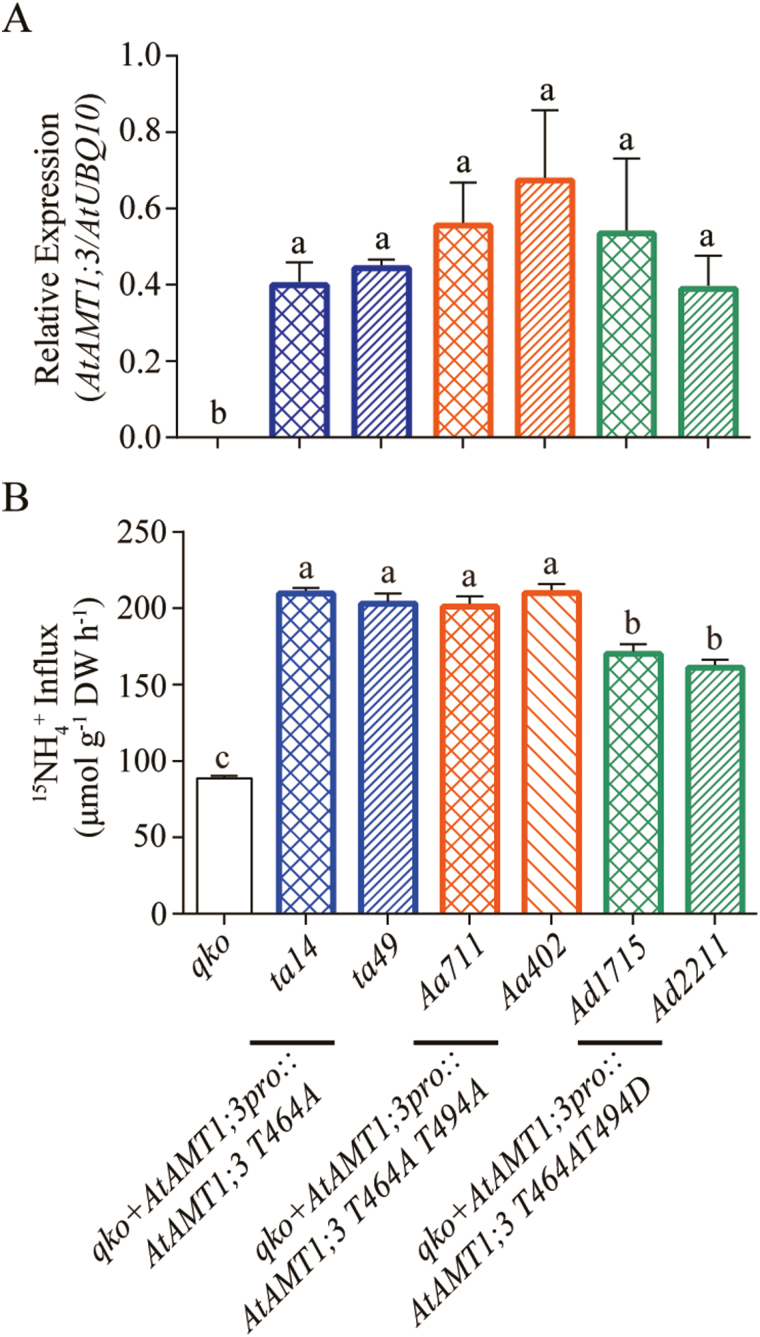
We then quantified the tuning ability of T494-dependent phosphorylation/dephosphorylation for root ammonium uptake when T464 was maintained in the non-phosphorylated status (Fig. 6). The transgenic lines qko+AtAMT1;3pro::AtAMT1;3 T464A (ta14 and ta49), qko+AtAMT1;3pro::AtAMT1;3 T464A T494A (Aa711 and Aa402) and qko+AtAMT1;3pro::AtAMT1;3 T464A T494D (Ad1715 and Ad2211) were generated with similar expression levels of AtAMT1;3 variants in roots (Fig. 6A). Quantification of the 15NH4+ influx rates in transgenic Arabidopsis roots expressing the double phospho-variant T464A T494A showed similar rates to those expressing the single phospho-variant T464A (Fig. 6B). By contrast, the lines expressing T464A T494D had approx. 32~34% reduced ammonium uptake activity compared with that of the non-phospho-mimic form T464A T494A. Thus, in addition to T464-mediated activation/inactivation, phospho-dependent modification of T494 adjusted ~30% of AtAMT1;3 transport activity in roots.
Fig. 6.
Functional characterization of AtAMT1;3 T464 T494 double phospho-mutants in transgenic Arabidopsis roots. (A) Transcript expression levels of AtAMT1;3 in roots of qko, qko+AtAMT1;3pro::AtAMT1;3 T464A, qko+AtAMT1;3pro::AtAMT1;3 T464A T494A or qko+AtAMT1;3pro::AtAMT1;3 T464A T494D transgenic lines. Levels were quantified by qPCR with three replicates and normalized by AtUBQ10 expression level. (B) Influx of 15N-labeled ammonium into roots of the same line as in (A). N-deficient roots were employed for 15N-labeled ammonium influx assays as determined at the concentration of 250 μM. Bars indicate means ±SD (n≥5) and significant differences at P<0.001 according to Tukey’s test are indicated by different letters. (This figure is available in color at JXB online.)
Discussion
Phosphorylation and dephosphorylation are flexible mechanisms for regulating the function of many nutrient transporters (including AKT1, NRT1;1, PHT1;1, and AMT1s) by reversibly altering their biochemical properties, subcellular location, or protein–protein interactions (Xu et al., 2006; Ho et al., 2009; Bayle et al., 2011; Straub et al., 2017). In many cases these transporters contain multiple phosphorylation sites, which probably provide a simple means for putting two or more such effects in the same protein for greatly increased regulatory potential (Cohen, 2000; Haruta et al., 2015). However, key questions remain to be addressed, including which sites are phospho-regulated under which biological conditions, as well as whether multisite phosphorylation events function independently or cooperatively to modulate transporter activity. In this study, among four multiple phosphorylation sites in the CTR of AtAMT1;3, we identified the T464 residue in the CTRC as inactivating transport activity via ammonium-induced phosphorylation. With the prerequisite of non-phosphorylated T464, a novel T494 residue was uncovered as fine-tuning transporter activity via nitrate-dependent phospho-regulation. These results provide a comprehensive model for how roots regulate nutrient transporter activity via multisite phosphorylation/dephosphorylation in response to various nutrient signals.
(De-)phosphorylation of T494 in the CTRNC fine-tunes AtAMT1;3 transporter activity with dependence on the CTRC
The cytosolic C-terminal domain, which is conserved between bacterial, fungal, and plant AMTs, has been demonstrated to serve as an allosteric regulator essential for regulating transport activity, as revealed by X-ray crystal structures and functional mutational analyses in vitro or in vivo (Loqué et al., 2007; Neuhäuser et al., 2007; Yuan et al., 2013; Boeckstaens et al., 2014; van den Berg et al., 2016). This domain is located in the conserved part of the CTR (CTRC) that includes the universally conserved ‘ExxGxD’ motif (Fig. 2A; Loqué et al., 2007; van den Berg et al., 2016). With the trimeric complex of AMTs, the interaction between the CTRC and cytosolic loops of its own monomer or those of the neighboring monomer controls the opening or closing of the transporters. In plant AMT1s, the CTRC undergoes significant conformational changes by phosphorylation of its conserved Thr, thereby breaking its interactions with internal cytosolic loops to inactivate the transporter (Fig. 7; Loqué et al., 2007; Neuhäuser et al., 2007; Yuan et al., 2013).
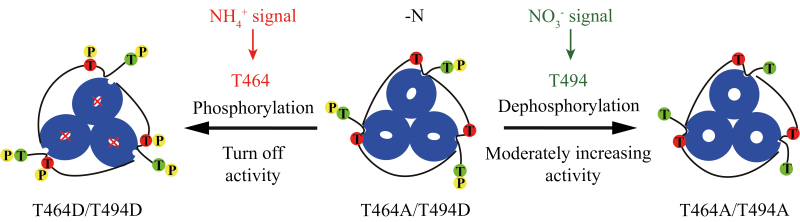
Fig. 7.
Schematic model for the regulation of AtATM1;3 transport activity via multisite phosphorylation/dephosphorylation in the C-terminal region. Two phosphorylation sites, T464 (labeled in red) in the CTRC and T494 (labeled in green) in the CTRNC, are involved in regulating AtAMT1;3 transport activity in response to ammonium and nitrate resupply, respectively. Under N deficiency (−N), AtAMT1;3 is in an active state with T464 non-phosphorylation (without yellow P) and T494 phosphorylation (with yellow P). This is mimicked by the T464A/T494D phospho-variant. Ammonium signals induce T464 phosphorylation (T464D/T494D) for shutting off the transporter activity via the allosteric trans-regulatory mechanism preventing excessive uptake of ammonium. Nitrate signals, however, transiently induce ammonium uptake by T494 dephosphorylation (T464A T494A) for moderate increase of transport activity. Non-phosphorylated T464 is a prerequisite of fine-tuning activity via T494 phosphorylation/dephosphorylation.
Several lines of evidence suggest a role of the CTRNC in regulating AMT transport activity. First, deletion of the CTRNC of AtAMT1;1 (Y469stop; Loqué et al., 2007) and AtAMT1;3 (H474stop; Supplementary Fig. S5) partially reduced their transporter activity. Second, phospho-dependent modifications at T494 located in the CTRNC of AtAMT1;3 allowed fine-tuning of transporter activity as shown in yeast, oocytes, and transgenic Arabidopsis roots (Figs 2–4). In addition, the CTRNC of the yeast MEP2 was dubbed an auto-inhibitory (AI) region, and deletion of the AI region or phosphorylation of the S457 residue at the AI region is able to activate the transporter function (Boeckstaens et al., 2014; van den Berg et al., 2016). Thus, besides the CTRC, the CTRNC can be involved in modulating AMT transporter activity, in either a positive or a negative manner.
The structural basis for MEP2 activation by S457 phosphorylation provides a clue for how modification of the CTRNC can affect the transporter activity (van den Berg et al., 2016). With S457 phosphorylation at the CTRNC, the CTRC may undergo a conformational change that is able to establish the interaction of CTRC with an inward-moving cytosolic loop for opening the transporter. Although it is unclear whether similar structural arrangements occur in plant AMTs, T494 phosphorylation in the CTRNC of AtAMT1;3 is expected to cause conformational changes of the CTRC. Unlike that generated by T464 phosphorylation to shut off transport activity completely, this conformational change of the CTRC via T494 only moderately reduced transport activity, without changing the affinity for ammonium (Figs 2–4). The CTRNC function in modulating AtAMT1;3 transporter activity relies on the CTRC as shown by the double phospho-variants in T464 and T494 (Figs 5 and 6; Supplementary Fig. S4). With the non-phospho-mimic T464A, T494 phosphorylation/dephosphorylation modulated function that is probably due to interactions between the CTRC and cytosolic loops for fine-tuning transporter activity. The regulatory function of T494, however, is completely lost when phosphorylation mimic T464D disrupts the interaction and closes the transporter.
Multiple phosphorylation sites, at least six Thr/Ser residues, are also present in the C-terminal tail of the Arabidopsis P-type H+-ATPase 2 (AHA2) proton pump (Rudashevskaya et al., 2012). Among them, phosphorylation at S899 and S931 decreases the activity, while phosphorylation at T881 or T947 increases the activity (Fuglsang et al., 2007; Takahashi et al., 2012; Haruta et al., 2014; Fuglsang et al., 2014). The phosphorylation at different sites may lead to distinct conformational changes of the C-terminus for these synergistic or antagonistic effects on AtAHA2 activity (Fuglsang et al., 2003). Multisite phosphorylation at the CTR of AtAMT1;3 revealed synergistic effects on the activities. The phosphorylation/dephosphorylation of T464 at the CTRC acted as a prime switch to turn on/off activity, while that of T494 at the CTRNC acted as a modulator to fine tune activity, allowing plants to precisely determine ammonium uptake capacity in response to diverse N conditions.
Multisite (de-)phosphorylation of AtAMT1;3 provides a mechanism for introducing controls of ammonium uptake by different N signaling pathways
Multisite phosphorylation can occur as phosphorylation at more than one residue by the same protein kinase, or phosphorylation of more than one site by two or more protein kinases (Cohen, 2000). For the latter, multisite phosphorylation is able to modulate the protein function by different kinases/phosphatases via different signaling pathways. To respond to various signals, multiple sites can be phosphorylated with different degrees and even with distinct directions of change for each site, providing a very flexible, but sophisticated, system for precisely regulating protein function (Haruta et al., 2015). For instance, AtAHA2 can be activated by phosphorylation of T947 via the auxin signaling pathway (Takahashi et al., 2012) or by phosphorylation of T881 by the receptor kinase tyrosine-sulfated peptide 1 receptor (PSY1R) via a peptide hormone signal (Fuglsang et al., 2014). This pump can be also inactivated by phosphorylation at other residues, i.e. S899, by receptor-like kinase FER protein kinase (FERONIA) via the peptide rapid alkalization factor (RALF) signal (Haruta et al., 2014) or at S931 by a Ser/Thr kinase PKS5 in response to stress (Fuglsang et al., 2007).
Similar to AtAHA2, the AtAMT1;3 transporter activity can be modulated by multisite phosphorylation under control of different signaling pathways (Figs 1 and 2). The T464 in the CTRC is phosphorylated after ammonium signals (Fig. 1). This ammonium-triggered Thr phosphorylation appears highly conserved for all plant AMT1s, i.e. T460 and T472 for AtAMT1;1 and AtAMT1;2, respectively (Straub et al., 2017), T464 for AtAMT1;3 (Fig. 1), and T453 for OsAMT1;2 (Beier et al., 2018). The kinase complex CIPK23–CBL1 phosphorylates AtAMT1;1 and AtAMT1;2, but not AtAMT1;3. Since OsACTPK1, an STY kinase, can phosphorylate the same residue in OsAMT1;2 (Beier et al, 2018), the corresponding Arabidopsis homologs might encode the candidate kinases for phosphorylating AtAMT1;3 at T464. More than one kinase is recruited in ammonium-triggered phosphorylation of conserved Thr in the CTRC, ensuring the inhibition of root ammonium uptake under elevated ammonium levels to prevent the risk of ammonium toxicity.
Nitrate signals can induce phosphorylation/dephosphorylation of AtAMT1;3 at several Thr or Ser residues in the CTRNC (Fig. 2; Supplementary Fig S2). The direction of phospho-dynamics for each site even differed under nitrate resupply. The T494 residue was transiently dephosphorylated, while S480 and S487 were transiently phosphorylated simultaneously (Fig. 2). The kinetics of changes in T494 phosphorylation status and the associated transporter activity closely paralleled those of ammonium uptake in roots, suggesting a regulatory role of T494 in the nitrate response (Figs 1 and 2). Unexpectedly, phosphorylation-mimicking mutations at either single or double sites of S480 and S487 did not lead to activity changes of AtAMT1;3 mutants in yeast (Fig. 2; Supplementary Fig. S6). The role of phosphorylation at S480 and S487, however, cannot be excluded because their phosphorylation might change subcellular localization, protein degradation, or interaction with other proteins in a plant-specific manner. If so, the yeast system is not suitable to investigate these effects. Nevertheless, mutations at these two residues had no direct effect on transport of ammonium. Unlike a single site phosphorylation control at the CTRC dominated by ammonium signals, multisite phosphorylation at CTRNC of AMT1s allows introduction of control by diverse N-related signaling pathways (i.e. nitrate, light, and hormone signals) (Supplementary Table S2). Indeed, multisite phosphorylation is required in the root epidermis-expressed AtAMT1;1 and AtAMT1;3, which allows precise regulation of the symplastic ammonium transport in response to multiple signals in dynamic environments. For the endodermis-expressed AtAMT1;2 that mediates the apoplastic transport pathway, by contrast, the presence of a single ThrCTR phosphorylation may be sufficient to shut off transport activity under high external ammonium, preventing excessive ammonium allocation to the more sensitive shoot.
Taken together, our data present a model for (de-)phosphorylation-based regulation of AtAMT1;3 at multiple phosphorylation sites under control of different N signals, which may be conserved in other plant AMT1s (Fig. 7). In N-deficient roots, AtAMT1;3 is present in an active form with non-phosphorylated T464 in the CTRC and phosphorylated T494 in the CTRNC (mimicked by T464A T494D). Ammonium signals rapidly induce T464 phosphorylation (T464D T494D) to shut off transport activity, preventing excessive uptake of ammonium. Nitrate signals, on the contrary, transiently induce ammonium uptake by T494 dephosphorylation (T464A T494A). This allows elevation of the activity of AtAMT1;3, which transports NH4+ as a counter ion to nitrate. The uptake is returned to the original level after T494 re-phosphorylation. Considering that ammonium and nitrate are a major cation and anion, respectively, in plants, the nitrate-trigged transient induction of ammonium uptake is probably relevant to change the uptake balance of both ions (Kronzucker et al., 1999; Babourina et al., 2007; Luo et al., 2013; Hachiya and Sakakibara, 2017). The CIPK23–CBL1 kinase complex is supposed to sense ammonium, nitrate, and potassium simultaneously for controlling cellular ion homeostasis (Xu et al., 2006; Ho et al., 2009; Straub et al., 2017). Given that many kinases and phosphatases are regulated via the nitrate signaling pathway (Undurraga et al., 2017), identification of those upstream modulators of AMT1s will be of great interest for further research.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Transcript expression levels of AtAMT1;3 in qko+13 roots in response to ammonium and nitrate resupply.
Fig. S2. Phosphorylation dynamics of AtAMT1;3 CTRNC at multiple sites (S480, S487, and T494) in response to nitrate or ammonium.
Fig. S3. Yeast growth complementation of AtAMT1;3 S480, S487, and T494 single phospho-mutants.
Fig. S4. Functional characterization of AtAMT1;3 T494 single phospho-mutants and T464 T494 double phospho-mutants in Xenopus oocytes.
Fig. S5. Yeast growth complementation of AtAMT1;3 CTRNC deletion mutant.
Fig. S6. Functional characterization of AtAMT1;3 T464 S480 S487 triple phospho-mutants in yeast.
Table S1. Primers used in this study.
Table S2. In vivo phosphorylation sites in the C-terminal region of AtAMT1s.
Acknowledgements
We thank the anonymous reviewers for their critical readings and constructive comments on the manuscript.
Glossary
Abbreviations:
- ACTPK1
ACT domain protein kinase 1
- AHA2
P-type H+-ATPase 2
- AMT
ammonium transporter
- AMT1
AMT1-type ammonium transporter
- CBL1
calcineurin B-like protein 1
- CIPK23
calcineurin B-like protein-interacting protein kinase 23;
- CTR
, C-terminal region
- CTRC
conserved part of the CTR
- CTRNC
non-conserved part of the CTR
- qPCR
quantitative RT-PCR
- STY
serine/threonine/tyrosine
- ThrCTR
the conserved Thr residue in the CTR
Funding
This work was financially supported by the National Natural Science Foundation of China (NSFC) [31430095; 31471934; 30971863; 30870189], and the Deutsche Forschungsgemeinschaft (DFG) [328017493/GRK 2366].
Author contributions
XW and LY conceived and designed the research. XW, TL, YZ, FD, and BN performed the experiments. XW, BN, UL, WS, and LY analysed the data. XW and LY wrote the manuscript and BN, UL, and WS helped to revise the manuscript. All authors read and approved the manuscript. The authors declare no conflict of interest.
References
- Babourina O, Voltchanskii K, McGann B, Newman I, Rengel Z. 2007. Nitrate supply affects ammonium transport in canola roots. Journal of Experimental Botany 58, 651–658. [DOI] [PubMed] [Google Scholar]
- Baukrowitz T, Tucker SJ, Schulte U, Benndorf K, Ruppersberg JP, Fakler B. 1999. Inward rectification in KATP channels: a pH switch in the pore. The EMBO Journal 18, 847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle V, Arrighi JF, Creff A, Nespoulous C, Vialaret J, Rossignol M, Gonzalez E, Paz-Ares J, Nussaume L. 2011. Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. The Plant Cell 23, 1523–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier MP, Obara M, Taniai A, et al. . 2018. Lack of ACTPK1, an STY kinase, enhances ammonium uptake and use, and promotes growth of rice seedlings under sufficient external ammonium. The Plant Journal 93, 992–1006. [DOI] [PubMed] [Google Scholar]
- Boeckstaens M, Llinares E, Van Vooren P, Marini AM. 2014. The TORC1 effector kinase Npr1 fine tunes the inherent activity of the Mep2 ammonium transport protein. Nature Communications 5, 3101. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. 2010. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464, 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cohen P. 2000. The regulation of protein function by multisite phosphorylation – a 25 year update. Trends in Biochemical Sciences 25, 596–601. [DOI] [PubMed] [Google Scholar]
- Duan F, Giehl RFH, Geldner N, Salt DE, von Wirén N. 2018. Root zone-specific localization of AMTs determines ammonium transport pathways and nitrogen allocation to shoots. PLoS Biology 16, e2006024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsberger WR, Schulze WX. 2012. Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. The Plant Journal 69, 978–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Borch J, Bych K, Jahn TP, Roepstorff P, Palmgren MG. 2003. The binding site for regulatory 14-3-3 protein in plant plasma membrane H+-ATPase involvement of a region promoting phosphorylation-independent interaction in addition to the phosphorylation-dependent C-terminal end. The Journal of Biological Chemistry 278, 42266–42272. [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, et al. . 2007. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. The Plant Cell 19, 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Kristensen A, Cuin TA, et al. . 2014. Receptor kinase-mediated control of primary active proton pumping at the plasma membrane. The Plant Journal 80, 951–964. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Lejay L, Gojon A, Ninnemann O, Frommer WB, von Wirén N. 1999. Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. The Plant Cell 11, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl RFH, Laginha AM, Duan F, Rentsch D, Yuan L, von Wirén N. 2017. A critical role of AMT2;1 in root-to-shoot translocation of ammonium in Arabidopsis. Molecular Plant 10, 1449–1460. [DOI] [PubMed] [Google Scholar]
- Gu R, Duan F, An X, Zhang F, von Wirén N, Yuan L. 2013. Characterization of AMT-mediated high-affinity ammonium uptake in roots of maize (Zea mays L.). Plant & Cell Physiology 54, 1515–1524. [DOI] [PubMed] [Google Scholar]
- Hachiya T, Sakakibara H. 2017. Interactions between nitrate and ammonium in their uptake, allocation, assimilation, and signaling in plants. Journal of Experimental Botany 68, 2501–2512. [DOI] [PubMed] [Google Scholar]
- Haruta M, Gray WM, Sussman MR. 2015. Regulation of the plasma membrane proton pump (H+-ATPase) by phosphorylation. Current Opinion in Plant Biology 28, 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. 2014. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. 2009. CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. [DOI] [PubMed] [Google Scholar]
- Kaiser BN, Rawat SR, Siddiqi MY, Masle J, Glass AD. 2002. Functional analysis of an Arabidopsis T-DNA “knockout” of the high-affinity NH4+ transporter AtAMT1;1. Plant Physiology 130, 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademi S, O’Connell J 3rd, Remis J, Robles-Colmenares Y, Miercke LJ, Stroud RM. 2004. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science 305, 1587–1594. [DOI] [PubMed] [Google Scholar]
- Kiba T, Inaba J, Kudo T, et al. . 2018. Repression of nitrogen starvation responses by members of the Arabidopsis GARP-type transcription factor NIGT1/HRS1 subfamily. The Plant Cell 30, 925–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronzucker HJ, Siddiqi MY, Glass AD, Kirk GJ. 1999. Nitrate-ammonium synergism in rice. A subcellular flux analysis. Plant Physiology 119, 1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V, Loqué D, Hörmann F, Yuan L, Bohner A, Engelsberger WR, Lalonde S, Schulze WX, von Wirén N, Frommer WB. 2009. Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. The Plant Cell 21, 3610–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Tian Y, Wu K, et al. . 2018. Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560, 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima JE, Kojima S, Takahashi H, von Wirén N. 2010. Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. The Plant Cell 22, 3621–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, von Wirén N. 2017. Ammonium as a signal for physiological and morphological responses in plants. Journal of Experimental Botany 68, 2581–2592. [DOI] [PubMed] [Google Scholar]
- Loqué D, Lalonde S, Looger LL, von Wirén N, Frommer WB. 2007. A cytosolic trans-activation domain essential for ammonium uptake. Nature 446, 195–198. [DOI] [PubMed] [Google Scholar]
- Loqué D, Mora SI, Andrade SL, Pantoja O, Frommer WB. 2009. Pore mutations in ammonium transporter AMT1 with increased electrogenic ammonium transport activity. The Journal of Biological Chemistry 284, 24988–24995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué D, von Wirén N. 2004. Regulatory levels for the transport of ammonium in plant roots. Journal of Experimental Botany 55, 1293–1305. [DOI] [PubMed] [Google Scholar]
- Loqué D, Yuan L, Kojima S, Gojon A, Wirth J, Gazzarrini S, Ishiyama K, Takahashi H, von Wirén N. 2006. Additive contribution of AMT1;1 and AMT1;3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient Arabidopsis roots. The Plant Journal 48, 522–534. [DOI] [PubMed] [Google Scholar]
- Luo J, Qin J, He F, Li H, Liu T, Polle A, Peng C, Luo ZB. 2013. Net fluxes of ammonium and nitrate in association with H+ fluxes in fine roots of Populus popularis. Planta 237, 919–931. [DOI] [PubMed] [Google Scholar]
- Marini AM, Soussi-Boudekou S, Vissers S, Andre B. 1997. A family of ammonium transporters in Saccharomyces cerevisiae. Molecular and Cellular Biology 17, 4282–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner P, Rengel Z. 2012. Nutrient availability in soils. In: Marschner P, ed. Marschner’s mineral nutrition of higher plants, 3rd edn San Diego: Academic Press, 315–330. [Google Scholar]
- Mayer M, Dynowski M, Ludewig U. 2006. Ammonium ion transport by the AMT/Rh homologue LeAMT1;1. The Biochemical Journal 396, 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M, Ludewig U. 2006. Role of AMT1;1 in NH4+ acquisition in Arabidopsis thaliana. Plant Biology 8, 522–528. [DOI] [PubMed] [Google Scholar]
- Menz J, Li Z, Schulze WX, Ludewig U. 2016. Early nitrogen-deprivation responses in Arabidopsis roots reveal distinct differences on transcriptome and (phospho-) proteome levels between nitrate and ammonium nutrition. The Plant Journal 88, 717–734. [DOI] [PubMed] [Google Scholar]
- Neuhäuser B, Dynowski M, Ludewig U. 2009. Channel-like NH3 flux by ammonium transporter AtAMT2. FEBS Letters 583, 2833–2838. [DOI] [PubMed] [Google Scholar]
- Neuhäuser B, Dynowski M, Mayer M, Ludewig U. 2007. Regulation of NH4+ transport by essential cross talk between AMT monomers through the carboxyl tails. Plant Physiology 143, 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat SR, Silim SN, Kronzucker HJ, Siddiqi MY, Glass AD. 1999. AtAMT1 gene expression and NH4+ uptake in roots of Arabidopsis thaliana: evidence for regulation by root glutamine levels. The Plant Journal 19, 143–152. [DOI] [PubMed] [Google Scholar]
- Rudashevskaya EL, Ye J, Jensen ON, Fuglsang AT, Palmgren MG. 2012. Phosphosite mapping of P-type plasma membrane H+-ATPase in homologous and heterologous environments. The Journal of Biological Chemistry 287, 4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda Y, Ikeda A, Saiki S, von Wirén N, Yamaya T, Yamaguchi J. 2003a Distinct expression and function of three ammonium transporter genes (OsAMT1;1–1;3) in rice. Plant & Cell Physiology 44, 726–734. [DOI] [PubMed] [Google Scholar]
- Sonoda Y, Ikeda A, Saiki S, Yamaya T, Yamaguchi J. 2003b Feedback regulation of the ammonium transporter gene family AMT1 by glutamine in rice. Plant & Cell Physiology 44, 1396–1402. [DOI] [PubMed] [Google Scholar]
- Straub T, Ludewig U, Neuhäuser B. 2017. The kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. The Plant Cell 29, 409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Hayashi K, Kinoshita T. 2012. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiology 159, 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Undurraga SF, Ibarra-Henríquez C, Fredes I, Álvarez JM, Gutiérrez RA. 2017. Nitrate signaling and early responses in Arabidopsis roots. Journal of Experimental Botany 68, 2541–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg B, Chembath A, Jefferies D, Basle A, Khalid S, Rutherford JC. 2016. Structural basis for Mep2 ammonium transceptor activation by phosphorylation. Nature Communications 7, 11337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wirén N, Gazzarrini S, Gojon A, Frommer WB. 2000. The molecular physiology of ammonium uptake and retrieval. Current Opinion in Plant Biology 3, 254–261. [PubMed] [Google Scholar]
- Wu X, Yang H, Qu C, Xu Z, Li W, Hao B, Yang C, Sun G, Liu G. 2015. Sequence and expression analysis of the AMT gene family in poplar. Frontiers in Plant Science 6, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH. 2006. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125, 1347–1360. [DOI] [PubMed] [Google Scholar]
- Xuan YH, Priatama RA, Huang J, et al. . 2013. Indeterminate domain 10 regulates ammonium-mediated gene expression in rice roots. New Phytologist 197, 791–804. [DOI] [PubMed] [Google Scholar]
- Yuan L, Gu R, Xuan Y, Smith-Valle E, Loqué D, Frommer WB, von Wirén N. 2013. Allosteric regulation of transport activity by heterotrimerization of Arabidopsis ammonium transporter complexes in vivo. The Plant Cell 25, 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Loqué D, Kojima S, Rauch S, Ishiyama K, Inoue E, Takahashi H, von Wirén N. 2007a The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. The Plant Cell 19, 2636–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Loqué D, Ye F, Frommer WB, von Wirén N. 2007b Nitrogen-dependent posttranscriptional regulation of the ammonium transporter AtAMT1;1. Plant Physiology 143, 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.