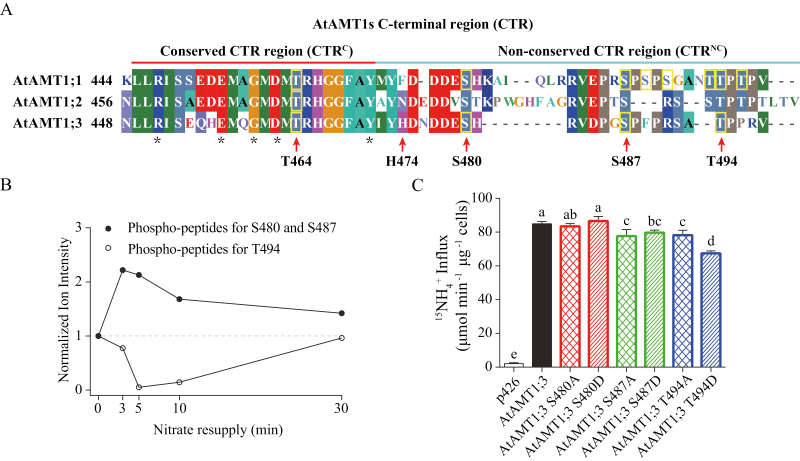
Fig. 2.
Phosphorylation dynamics of AtAMT1;3 CTRNC at multiple sites (S480, S487, and T494) in response to nitrate and functional analysis of corresponding phospho-mutants in yeast. (A) In vivo phosphorylation sites in the CTR of AtAMT1s identified by phosphoproteomics and phospho-specific antibodies (Supplementary Table S1). Alignment of amino acid sequences showed that the CTR of AtAMT1s consisted of a conserved (CTRC) and a non-conserved (CTRNC) part that are underlined in red and blue, respectively. The phosphorylated Thr/Ser residues are labeled with a yellow box. Five universally conserved residues within the CTRC, including the ‘ExxGxD’ motif, are labeled with asterisks. (B) Normalized ion intensity of phosphorylated AtAMT1;3 peptides HGGFAYIYHDNDDES(ph)HRVDPGS(ph)PFPR (S480/S487) and SAT(ph)PPRV (T494) that responded to nitrate. The data were obtained from Engelsberger and Schulze (2012). (C) Influx of 15N-labeled ammonium into yeast mutant 31019b expressing empty vector p426, AtAMT1;3 wild type, and different AtAMT1;3 phospho-variants: S480A, S480D, S487A, S487D, T494A, and T494D. Influx assays were performed at the concentration of 250 μM. Bars indicate means ±SD (n=5) and significant differences at P<0.001 according to Tukey’s test are indicated by different letters.