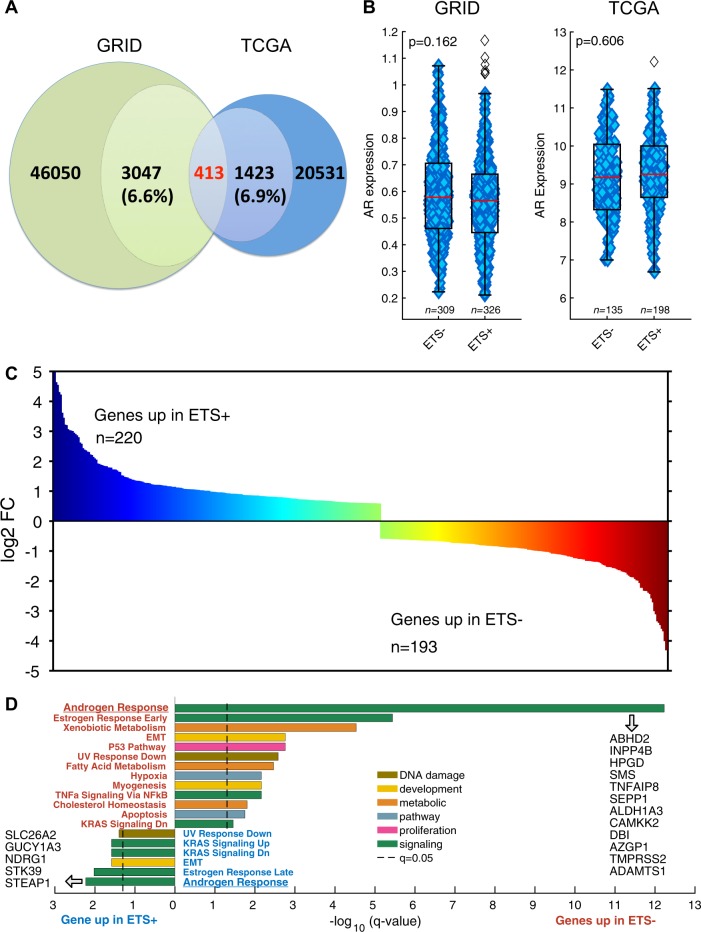
Fig. 2.
Genes differentially expressed in ETS+ versus ETS− PCa tumors. a The GRID dataset has 46050 genes with 3047 (6.6%) being differentially expressed when comparing ETS+ vs. ETS− PCa. The TCGA dataset has 20531 genes with 1423 (6.9%) being differentially expressed based on ETS status. Using a false-discovery-rate adjusted (q < 0.05) Mann–Whitney U test and a fold-change cut-off of 0.585 (TCGA) and 0.05 (GRID), 413 differentially expressed genes based on ETS status were defined in both PCa databases. b There is no significant difference in AR expression between ETS− and ETS+ tumors in either TCGA or GRID. The 413 significant differentially expressed genes (c) in the TCGA and GRID PCa based on ETS status were analyzed by GSEA and the HALLMARK gene sets [43]. Significant gene sets (d) overexpressed in ETS+ are shown at the top and those gene sets for ETS− shown below. Androgen response was the top-ranked gene set for both ETS+ and ETS− significantly overexpressed genes