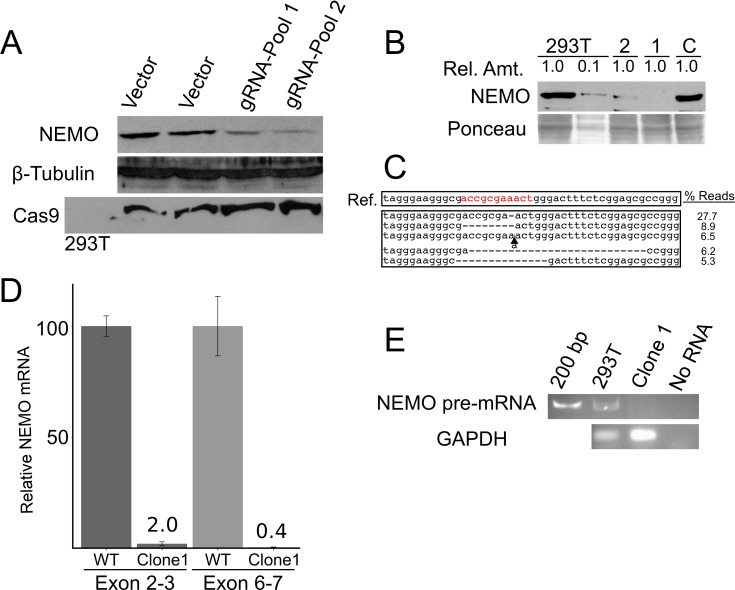
Fig 2. CRISPR/Cas9-based targeting of the exon 1b core promoter disrupts NEMO protein expression in 293T cells.
(A) 293T cells that were infected with LentiCRISPR2.0 construct containing no gRNA or the exon 1B gRNA (gRNA-Pool 1 and -2) were selected with puromycin, and puromycin-resistant pools of cells were then subjected to Western blotting for NEMO, β-tubulin (as a loading control), or FLAG-Cas9. (B) Shown is an anti-NEMO Western blot of control 293T cells, a 1:10 dilution of the control extract, and two clones (1 and 2) derived from 293T cells transduced with the LentiCRISPR-exon 1B virus. A Ponceau stain of the filter (as a loading control) is shown at the bottom. As a further control, a clone of puromycin-resistant cells targeted with a different gRNA were analyzed (C). (C) Sequencing of the targeted genomic locus in clone 1 cells was done by PCR amplification of the targeted site and Illumina-sequencing of the PCR product. Shown at the top is the wild-type reference sequence, and below that, the five most abundant disruptions and their read frequencies among the ~34,000 total genomic sequence reads. The complete array of deletions and their frequencies are shown in Fig A in S1 File. (D) qPCR of NEMO transcripts (using exon 2 and 3 or exon 6 and 7 primers) was performed with RNA from control cells and clone 1 cells. The amount of NEMO mRNA is relative to the amount of RNA in control 293T cells (100). (E) RT-PCR using primers in exon 1B and the flanking intron 1 was performed with RNA from control and clone 1 cells. Products were then analyzed by gel electrophoresis and staining with ethidium bromide. As a control, shown also is an RT-PCR for GAPDH mRNA.