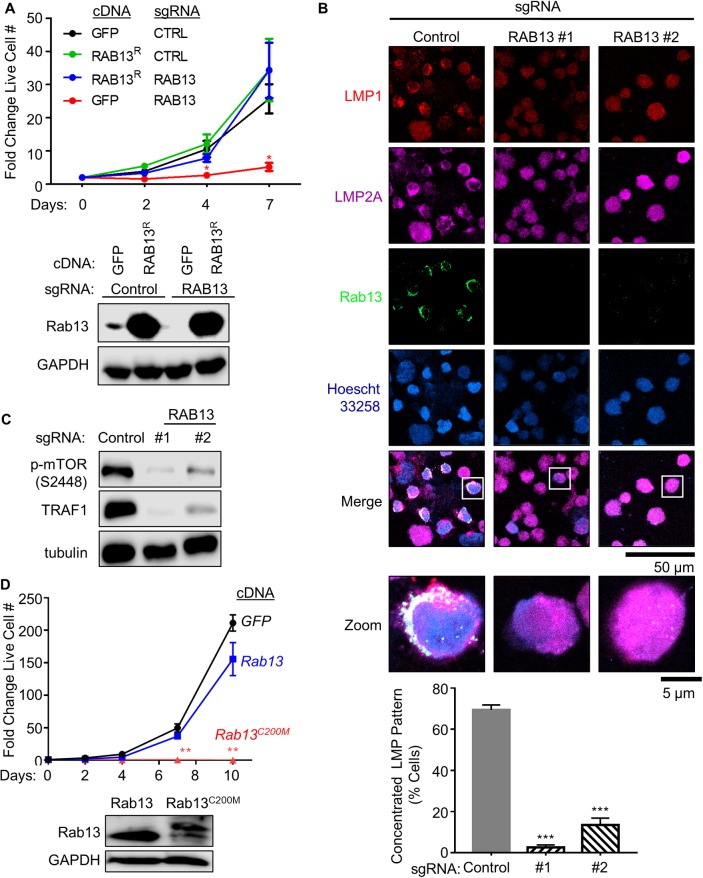
Fig 7. EBV-induced Rab13 is important for LCL LMP1 and 2A trafficking and signaling.
(A) Growth curve analysis of Cas9+ GM12878 LCL that express either control GFP or rescue RAB13 (RAB13R) cDNAs and either control or RAB13-targeting sgRNA. A silent mutation at the CRISPR PAM site abrogates Cas9 cleavage of the RAB13R cDNA. Data show the mean + SEM fold change live cell values at the indicated timepoints for n = 3 replicates. *, p<0.05. Below, representative immunoblots from n = 3 replicates of WCL from LCLs expressing the indicated cDNA and sgRNA are shown. (B) Immunofluorescence micrographs of Cas9+ GM12878 LCL that express either control or RAB13-targeting sgRNAs, showing localization patterns of Rab13, LMP1 or LMP2A. Merged images are shown with white boxes indicating inset images that have been further magnified. Scale bars are indicated (50 μm for single-channel and merged images; 5 μm for inset images). Shown below are mean + SEM numbers of cells with concentrated LMP1 and 2A staining patterns in GM12878 expressing control versus RAB13 targeting sgRNAs, for which 200 cells were counted from n = 3 replicates. (C) Immunoblot analysis of WCL from Cas9+ GM12878 LCL that express either control or independent RAB13-targeting sgRNAs. Representative blots from n = 2 experiments are shown. mTOR phosphoSerine-2448 (S2448) is a marker of mTOR activation, whereas TRAF1 is an LMP1-induced LCL target gene. See also S10 Fig. (D) Growth curve analysis of GM12878 LCL that stably express either control GFP, wildtype Rab13 or a Rab13 point mutant in which the putative cystine 200 prenylation site has been changed to methionine to abrogate GGPP modification (Rab13C200M). Data show the mean + SEM fold change live cell values at the indicated timepoints for n = 3 replicates. Below, representative immunoblots from n = 3 replicates of WCL from LCLs expressing the indicated RAB13 cDNA are shown.