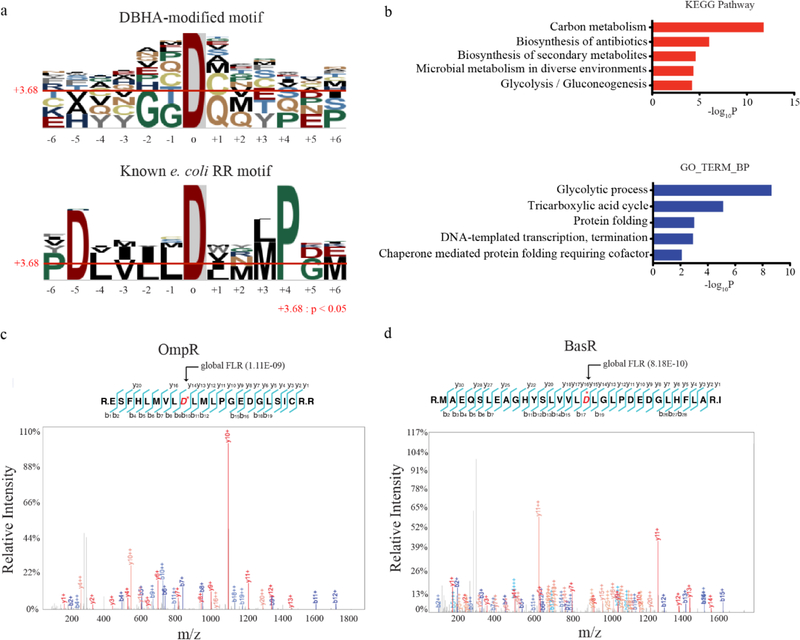
Figure 2.
a) Enriched sequence motifs derived from DBHA-modified aspartic acid sites (top) compared to the motif generated by known response regulator pD sites in E. coli (bottom). b) Gene-ontology biological process categories (GOTERM BP; bottom) and KEGG pathways (top) enriched among DBHA-modified proteins. Representative, statistically significant categories are shown, with a complete list in Figure S4. c,d) MS/MS spectra of DBHA-modified tryptic peptides at two known pD sites in the response regulator proteins c) OmpR and d) BasR. Observed b- and y-ions are labelled on each spectrum and highlighted on the tryptic peptide. The known pD sites at D55 and D51 in OmpR and BasR, respectively, are highlighted and starred (*) in red.