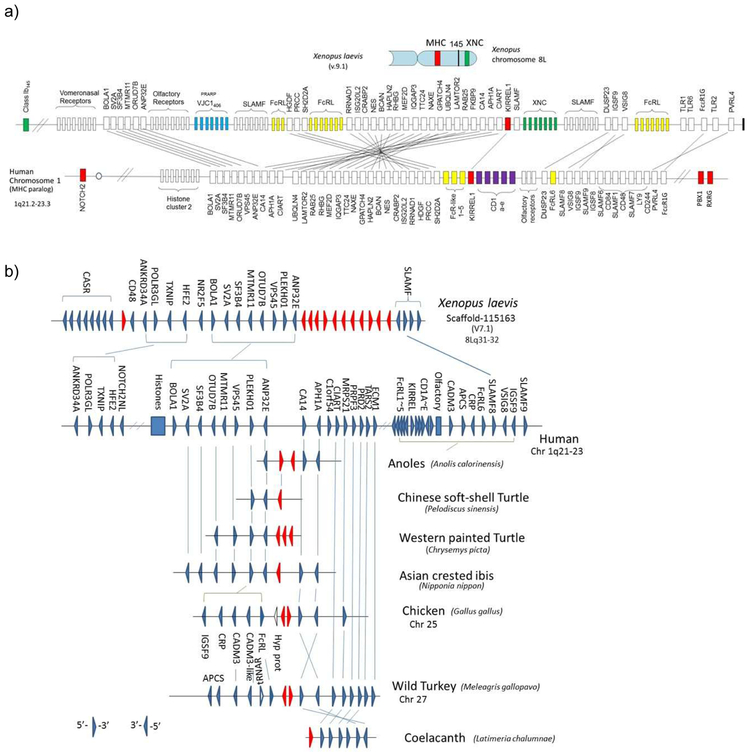
Figure 3. Human chromosome 1q21.2-23.3 is likely a translocated MHCpara.
a) Comparative mapping of the Xenopus XNC region (top) and human chromosome 1q21-23 region (bottom). The gene cluster, including CD1 (purple boxes), mapping to the human chromosome 1q21-23 region has its counterparts in the Xenopus XNC region. The XNC maps to the telomeric region of the Xenopus MHC chromosome and this region was proposed to be the result of translocation from the MHC. Other immune genes, such as slamf and fcr-like, are also found in this linkage group, suggesting the ancient linkage of these genes to the PIC (also refer to Fig 5a). Furthermore, the presence of uninterrupted IgL-like (VJC1406) genes (shown in blue boxes) provides a strong case for ancestral MHC-AgR linkage. Marker genes as well such as KIRREL are shown in red boxes. Only relevant genes are shown in this figure and the complete list of X. laevis genes is provided in the Suppl Table. The solid bar on the far right end of the Xenopus chromosome indicates the telomere.
b) Conserved synteny of novel AgR-like VJC1406 (PRARP) genes among frog, birds, and reptiles.
VJC1406 genes, most related to IgL genes, were found in other vertebrate species besides Xenopus and synteny is well conserved. Triangles indicate the 5’ to 3’ gene orientation. Red triangles represent AgR IgL-like genes, VJC1406. The number of genes varies depending on the species, and these loci have been lost in humans and bony fish. While the genes are present in cartilaginous fish, there is no information on synteny. Synteny is consistent with previously published data (73), however we focused more on the context of particular genes found in MHCtrans.