Figure 10. HHcy+T2DM induced Ly6C expression possibly via CEBPα induction.
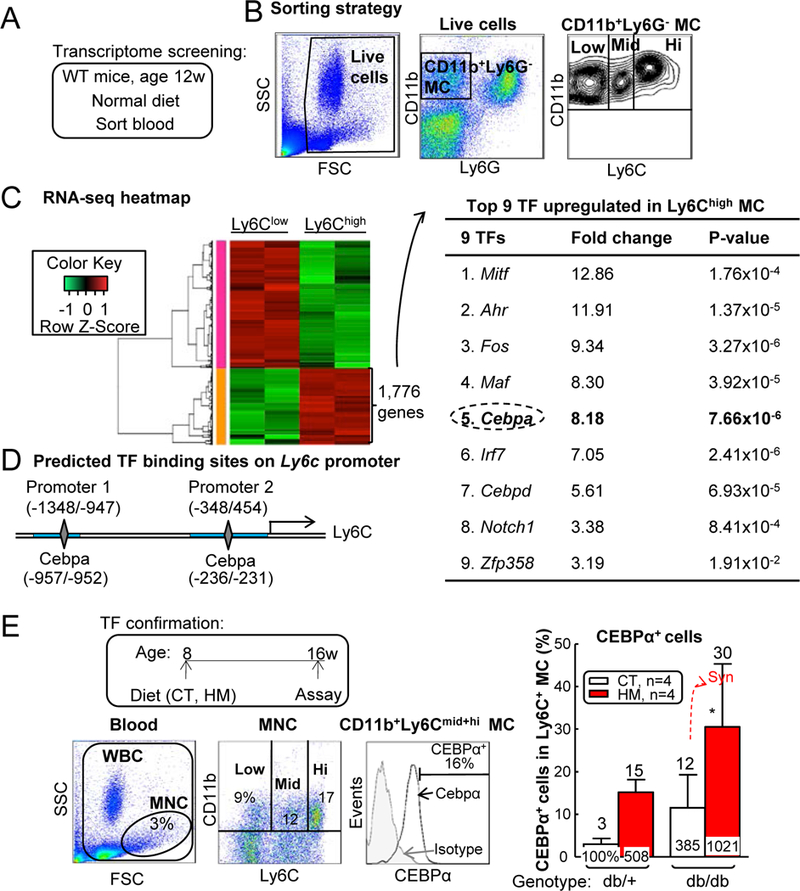
(A) Schematic illustration of transcriptome screening. (B) Sorting strategy for CD11b+Ly6G-Ly6Chigh inflammatory MCs (Ly6Chigh) and CD11b+Ly6G-Ly6Clow resident MCs (Ly6Clow) in WT mouse blood. Each population obtained 200,000 cells and RNA was extracted for RNA sequencing. (C) Upregulated TFs in Ly6Chigh cells. Using HiSeq X Ten system, we obtained 30 million reads from each sample, among which 17,230 genes were recognized. Ly6Chigh population had 1,776 genes upregulated compared with Ly6Clow group. Nine upregulated transcription factors were identified within Ly6Chigh 1,776 gene list. (D) Ly6c promoter analysis. Two core promoter regions (−1348/−947, −348/454, blue rectangle) were predicted by Encode. CEBPα transcription factor binding sites (−957/−952, −236/−231, grey diamond) were identified in mouse Ly6c core promoter regions by PROMO. (E) Frequency of CEBPα+ cells among total Ly6C+ MCs. N=4 mice. *P<0.05 vs CT diet mice with the same genotype. Synergy was defined as HHcy and T2DM produced a greater effects in db/db mice on HM diet than the sum of that in HHcy and T2DM alone. WBC, white blood cell.
