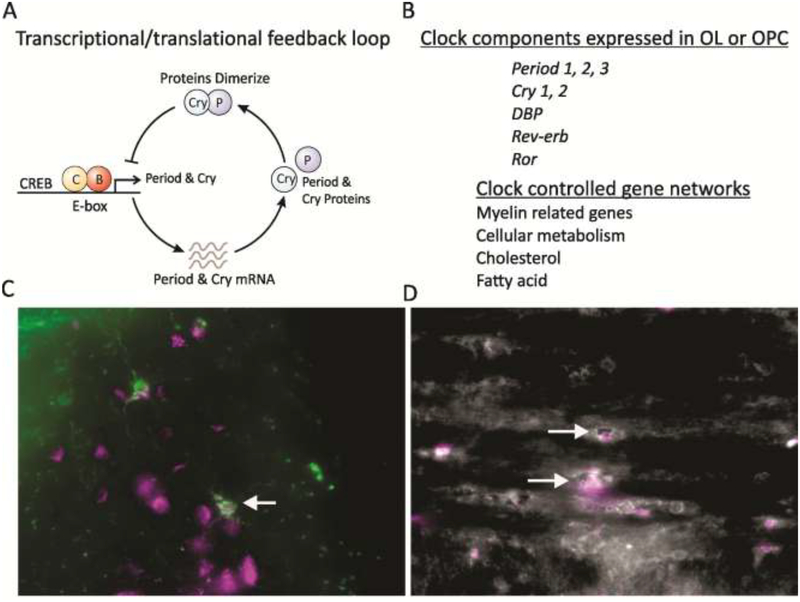
Fig. 2: Circadian timing system is likely to be active in OPC and OL.
A. schematic of the transcriptional/translational negative feedback loop that drives circadian rhythms in gene expression in most cells in our body. At the beginning of the cycle, CLOCK and BMAL1 protein complexes bind a specific promoter region (E-box) to activate the transcription of a family of genes including the Period (Per1/Per2/Per3) and Cryptochrome (Cry1/Cry2) genes. The levels of the transcripts for Per and Cry reach their peak during mid to late day, while the PER and CRY proteins peak in the early night. The PERs, CRYs, and other proteins form complexes in the cytoplasm that translocate back into the nucleus and turn off the transcriptional activity driven by CLOCK–BMAL1 with a delay (due to transcription, translation, dimerization, nuclear entry). The proteins are then degraded by ubiquitation allowing the cycle to begin again. In its simplest form, many cells contain this molecular feedback loop that regulates the rhythmic transcription of a number of genes. Additional feedback loops serve to contribute to the precision and robustness of this core oscillation. B. Microarray analysis indicates that OL express most of the genes that generate circadian oscillations [47]. The temporal profile of clock gene expression in OLs has not been established. A number of gene networks critical to OL function are known to be rhythmic and listed in this figure. C. O4 (left, green) and D. CNPase (right, white) positive OL in the white matter of adult C57bl/6j mice express PER2 (magenta). Arrows highlight OL co-expressing the markers. Mice were perfused at Zeitgeber Time (ZT) 6 and double-immunolabelling for O4 or CNPase and Per2 was performed as previously reported [158,159]. PER2 expression can be also appreciated in other neural cells surrounding the O4 positive OL. It should be noted that cells from different lineages will exhibit the peak of PER2 expression at different phases of the daily rhythm. The O4 hydridoma was a kind gift of Drs. Pfeiffer and Bansal, University of Connecticut [160,161].