Fig. 6.
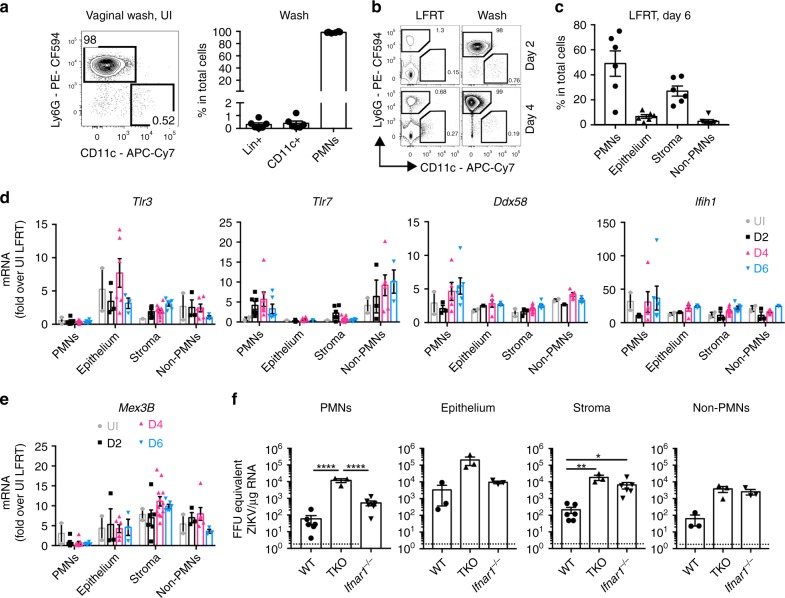
Compartmentalized role of viral sensing and IFNAR signaling in the LFRT. a Cells from the vaginal lumen of DMPA-treated uninfected C57BL/6N female mice were analyzed by flow cytometry. In the vaginal wash, frequencies of lineage positive (Lin+: CD19+ TCRβ+ NK1.1+), DCs (CD11c+: CD19− TCRβ− NK1.1− IAIE+ CD11c+), and polymorphonuclear neutrophils (PMNs: CD19− TCRβ− NK1.1− Ly6G+ CD11b+ SSC−) are also shown. Data represent one of four independent experiments with similar results, n = 6 mice. b Representative flow cytometry plots showing the abundance of Ly6G+ cells in the vaginal lumen (wash) compared with the LFRT tissue, at days 2 and 4 post vaginal infection with ZIKV. c Frequency of the four different sorted cell populations among total cells; See Supplementary Fig. 3 for sorting strategy. d, e Indicated genes were detected by qRT-PCR in total RNA from the sorted cell populations; UI n = 4 (sorted cells from two mice were combined for each data point), D2 and D6 n = 6 mice per time-point (sorted cells from two mice were combined for each data point in epithelium and non-PMNs due to low frequencies), and D4 n = 6 mice. f ZIKV RNA was detected by qRT-PCR in total RNA from the sorted cell populations; n = 6 mice per genotype. Due to low frequencies of epithelial cells and non-PMNs, cells from two different mice from WT or Ifnar1−/− mice were combined during sorting for subsequent RNA extraction and qRT-PCR. For TKO mice, cells in each population from two mice were combined together for RNA extraction. Data represent one of two independent experiments with similar results; mean ± SEM; *p < 0.05, **p < 0.01, ****p < 0.0001; one-way ANOVA with Tukey’s multiple comparison test. UI, uninfected. Source data for Fig. 5a, c–f are provided as a Source Data file