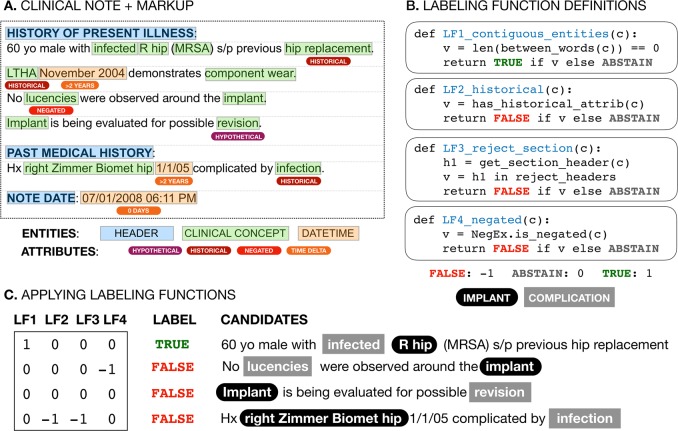
Fig. 6.
Labeling function examples. Clinical notes (top left) are preprocessed to generate document markup, tagging entities with clinical concepts (blue), parent section headers (green), dates/times (orange), and historical, hypothetical, negated, and time-delta attributes (shades of red). Labeling functions (top right) use this markup to represent domain insights as simple Python functions, e.g., a function to label mentions found in “Past Medical History” as FALSE because they are likely to be historical mentions rather than a current condition. These labeling functions vote {FALSE, ABSTAIN, TRUE} on candidate relationships (bottom, with true labels indicated in the LABEL column) consisting of implant (black rounded box) - complication (grey square box) pairs to generate a vector of noisy votes for each candidate relationship. The data programming step described in Fig. 1 uses this labeling function voting matrix to learn a generative model which assigns a single probabilistic label to each unlabeled candidate relationship