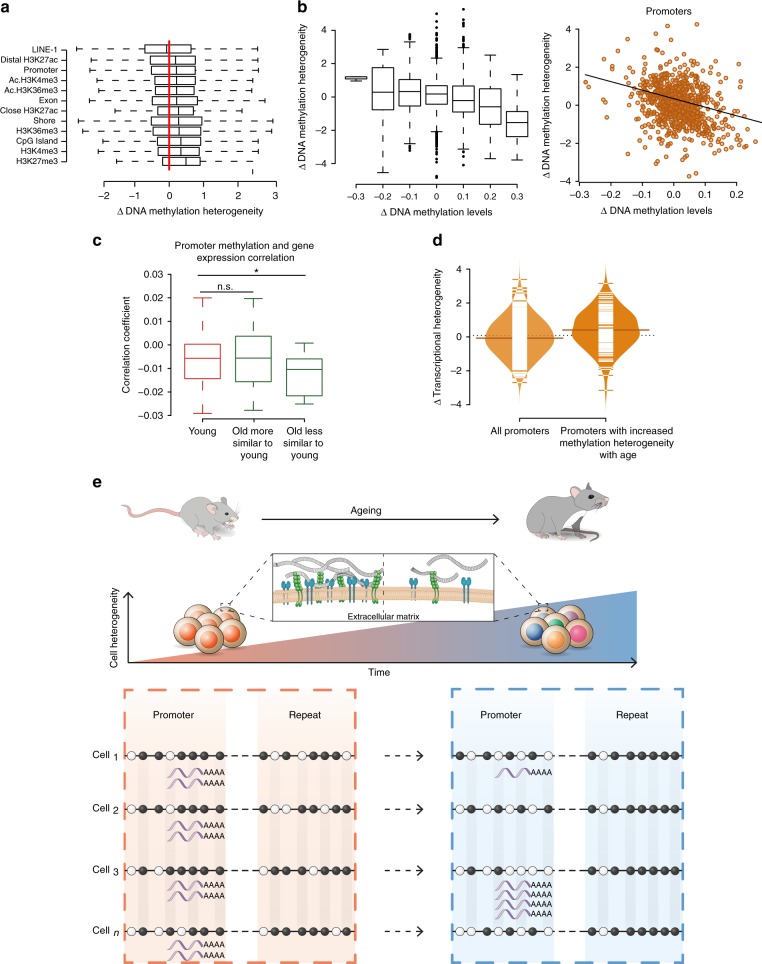
Fig. 4.
Changes in cell-to-cell methylation heterogeneity during ageing. a Normalised methylation heterogeneity changes with age (Δ methylation heterogeneity: old–young) across different genomic features (Chip-seq data from 2-months-old mice20,27, Ac: activated muscle stem cells). For all boxplots, the box represents the interquartile range and the horizontal line in the box represents the median. b Genome-wide normalised methylation heterogeneity difference with ages (Δ methylation heterogeneity: old–young) binned by 0.1 methylation level differences (left). Changes in promoter methylation heterogeneity (y-axis) and methylation levels (x-axis) with age (right). For all boxplots, the box represents the interquartile range and the horizontal line in the box represents the median. Source data are provided in Supplementary Data 4 (n = 2,355). c Distribution of Pearson’s correlation coefficients between promoter DNA methylation and gene expression (one association test per cell, number of cells: young = 64, old more similar to young = 30, old less similar to young = 20, *P < 0.05). For all boxplots, the box represents the interquartile range and the horizontal line in the box represents the median. d Increase of transcriptional heterogeneity with age across all promoters (n = 394) and promoters with increased DNA methylation heterogeneity (Δ methylation heterogeneity > 0.3, n = 113) (P < 0.001). Source data are provided in Supplementary Data 4. e Global increase of transcriptional cell-to-cell variability with age with enhanced heterogeneity in the multiple extracellular matrix-related genes (top). Relationship between transcriptional and DNA methylation heterogeneity in aged muscle stem cells (bottom). Empty circles represent unmethylated CpG sites and filled circles methylated CpG sites. Repeat elements become more homogeneous with age by increasing their methylation levels in a coordinated manner. In contrast, promoter regions become more heterogeneous by randomly losing DNA methylation, and this is coupled with an increase in transcriptional variability of the genes they drive