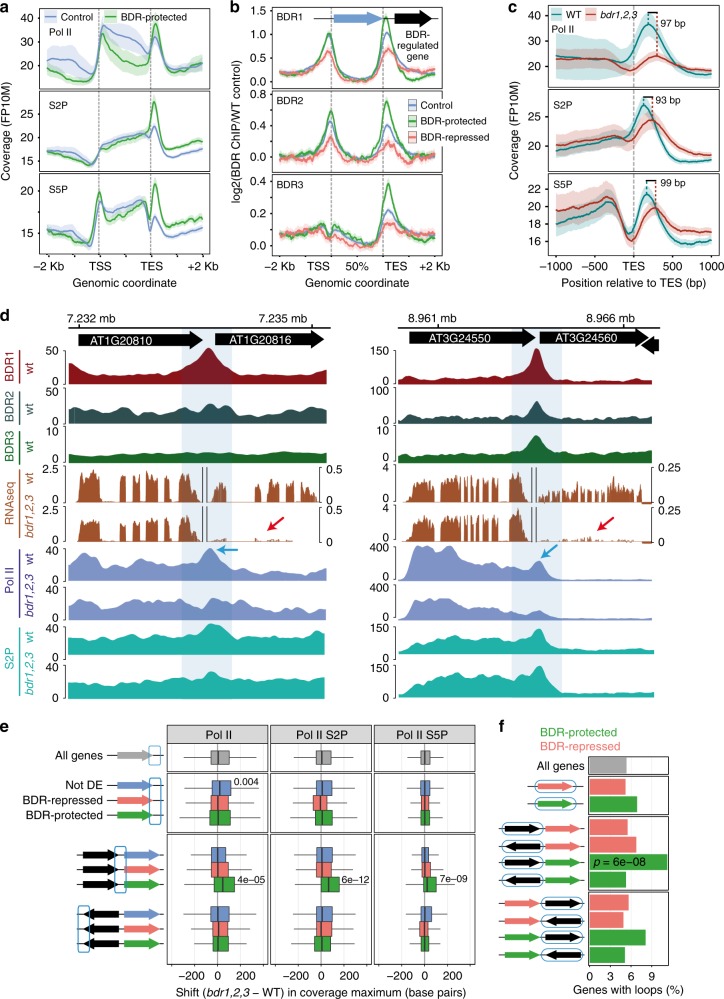
Fig. 6.
BDR proteins protect downstream genes from TI. a Levels of 3′ paused Pol II are elevated at tandem upstream neighbors of BDR-protected genes compared to expression-matched control genes. Metagene profiles of Pol II, Pol II-S5P, and Pol II-S2P ChIP-seq coverage across expressed genes in Arabidopsis seedlings. b BDR1 and BDR2 are enriched in the intergenic region between BDR-protected genes and their upstream tandem neighbors. Metagene profiles of ChIP-seq coverage at genes located upstream, on the same strand, as BDR-protected, BDR-repressed, or expression-matched control genes. c 3′ paused Pol II at upstream genes is reduced and shifted downstream in the absence of BDR proteins. Pol II near the TES of tandem upstream neighbors of BDR-protected genes is shifted ~96 bp downstream in the bdr1,2,3 mutant. Average Pol II ChIP-seq profiles are presented. d Browser tracks of BDR-protected genes and their upstream tandem neighbors. Note the high BDR1 and BDR2 occupancy in the intergenic region, reduction in the expression of the downstream gene in the bdr1,2,3 mutant (red arrows), and the reduction in 3′ paused Pol II at the upstream gene (blue arrows). e The downstream shift in the position of 3′ pausing in bdr1,2,3 occurs preferentially at the upstream tandem neighbors of BDR-protected genes. The centerline of boxplots is the median. The bounds of the box are the first and third quartiles (Q1 and Q3). Whiskers represent data range but are bounded to 1.5-fold the interquartile range (Q3-Q1); points outside this range are omitted. f Upstream tandem neighbors of BDR-protected genes are enriched in gene loops36. Enrichment of gene loops in BDR-regulated genes and their neighbors. Statistics reflect the presence of loops in the circled gene in each context. p Values are shown for Fisher exact test with BH p value correction