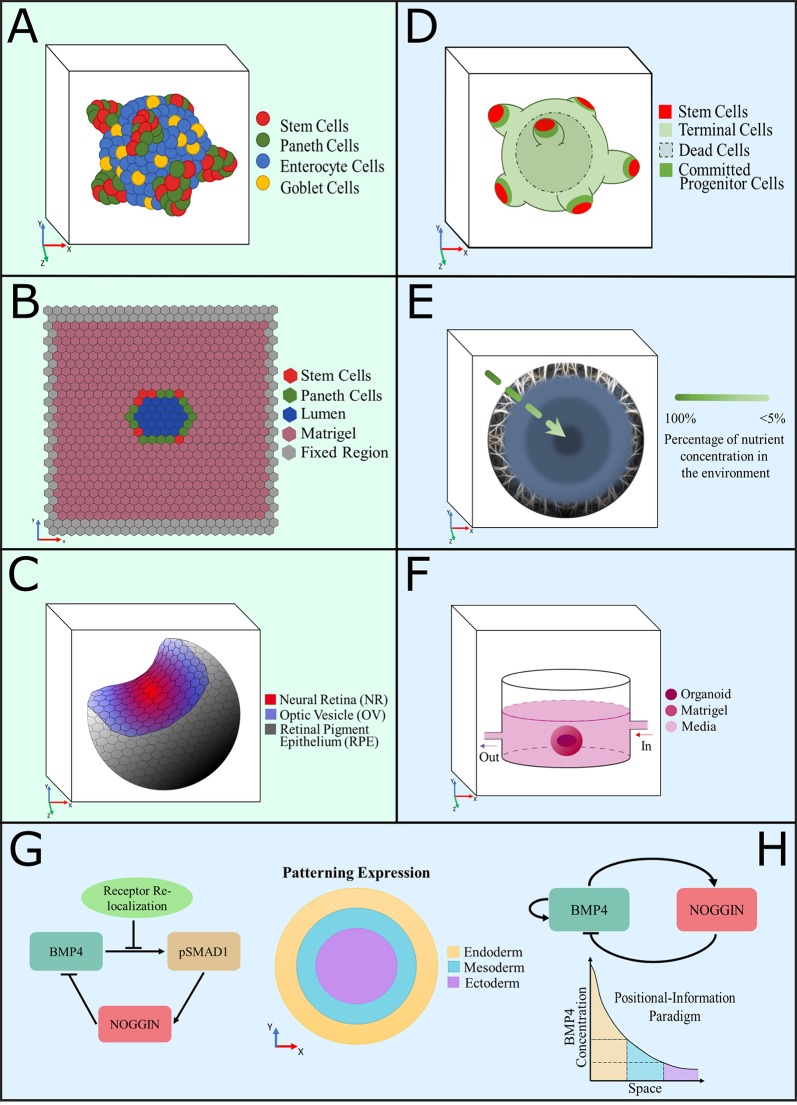
Figure 1.
Graphical representation of computational models developed to understand the intrinsic dynamics of organoid cultures. (A–C) Agent-based models, (D–H) equation-based models, color coded as per description in each panel. (A) A 3D model of intestinal organoids developed to investigate the distribution of cell populations and growth patterns provoked by Wnt and Notch signaling dynamics (Buske et al., 2012; Thalheim et al., 2018). (B) A 2D model of the cross section of a confluent intestinal epithelial layer, designed to study the biomechanical interactions between cells to produce crypt fission (Langlands et al., 2016; Almet et al., 2018). (C) Representation of a simulated optic-cup organoid (Okuda et al., 2018a). (D) Computational model of colon organoids created to study the effect of exogenous substances in the growth pattern and spatial distributions, to compare them with cancer phenotypes (Yan et al., 2018). (E) Diffusion model of a spheroid that simulates the consumption of nutrients in cerebral organoids to predict growth patterns (McMurtrey, 2016). (F) Model of oxygen consumption by a midbrain organoid grown in a millifluidic chamber to compare it with the oxygen consumption that occurred in the common well (Berger et al., 2018). (G, H) Equation-based reaction-diffusion models of gene networks used to simulate and predict fate patterning expression in gastruloids. The patterning expression and positional information paradigm plots show the signaling expressions of each primary germ layer observed experimentally (Etoc et al., 2016; Tewary et al., 2017).