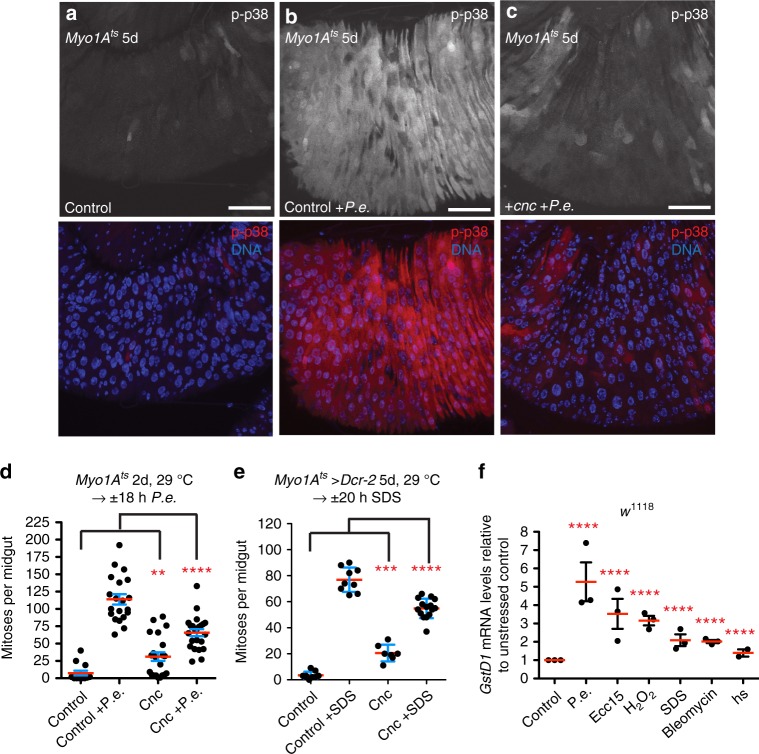
Fig. 6.
ROS-p38 signaling in ECs promotes ISC proliferation upon damage. a–c ROS are required for p38 activation in ECs upon P.e. infection. p38 activation increases in ECs in P.e.-infected control midguts (EC-specific, Myo1Ats) (b below, red) compared with uninfected control midguts (a below, red). p38 activation is blocked, however, in P.e.-infected midguts overexpressing Nrf2 (Cnc) in ECs for 5 days with Myo1Ats (c below, red) compared with P.e.-infected control midguts. d, e Nrf2 overexpression in ECs blocks ISC-mediated regeneration upon P.e. infection and detergent exposure. The ISC response is blocked in P.e.-infected midguts overexpressing Nrf2 in ECs for 2 days with Myo1Ats compared with P.e.-infected control midguts (Mann–Whitney; p < 0.0001). ISC mitoses are increased in midguts overexpressing Nrf2 for 2 days compared with control midguts (Mann–Whitney; p = 0.0011). The mean number of phosphorylated histone H3 Ser 10-positive cells per midgut with s.e.m. in control midguts (Myo1Ats) (n = 13 midguts), P.e.-infected control midguts (n = 20 midguts), midguts expressing Nrf2 (cnc) in ECs for 2 days with Myo1Ats (n = 22 midguts) and in P.e.-infected midguts expressing cnc in ECs for 2 days (n = 23 midguts). Midguts pooled from two independent experiments. d ISC-mediated regeneration is blocked in SDS-fed (20 h) midguts overexpressing Nrf2 (Cnc) in ECs for 6 days with Myo1Ats compared with SDS-fed control midguts (unpaired t; p < 0.0001). ISC mitoses are increased in midguts overexpressing Nrf2 for 6 days compared with control midguts (unpaired t; p < 0.0001). The mean number of phosphorylated histone H3 Ser 10-positive cells per midgut with s.e.m. from one representative experiment in control midguts (Myo1Ats > Dcr-2) (n = 9 midguts), SDS-fed control midguts (n = 9 midguts), midguts expressing Nrf2 (Cnc) in ECs for 6 days with Myo1Ats (n = 7 midguts) and in SDS-fed midguts expressing Cnc in ECs for 6 days (n = 15 midguts) (e). f Multiple stresses induce ROS-Nrf2 target gene GstD1 expression. Mean relative GstD1 mRNA levels with s.e.m. from n = 3 independent experiments (one-way ANOVA: p = 0.0086; Dunnett’s: P.e. < 0.0001, Ecc15 < 0.0001, H2O2 < 0.0001, SDS < 0.0001, bleomycin < 0.0001, heat shock < 0.0001) determined by qPCR from stressed and unstressed w1118 midguts. DNA is in blue. Scale bars in a–c are 50 μm. Representative images in a–c from three experiments. Source data are provided as a Source Data File