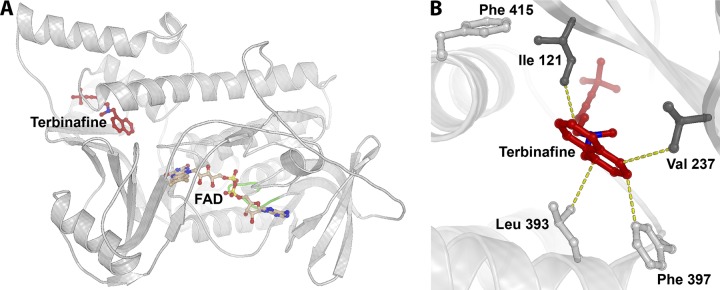
FIG 1.
Trichophyton rubrum squalene epoxidase model. (A) Overall view of protein fold with the cofactor FAD (light brown) superimposed from the structurally homologous flavoprotein/dehydrogenase from Cytophaga hutchinsonii (PDB entry 3NIX). The coordinates for terbinafine (red) are superimposed from the model of Saccharomyces cerevisiae SE (28), followed by refinement of the binding site positioning using AutoDock Vina. The loop comprising the conserved 45GXGXXG50 motif of the Rossmann fold is marked in green. (B) Close-up of the binding site of terbinafine. Amino acid positions where substitutions have been shown to be crucial for terbinafine resistance are depicted as sticks. The dark gray sticks represent the positions of newly discovered resistance mutations (I121 and V237), while the light gray sticks represent those identified previously (L393, F397, and F415) (2). Terbinafine is shown in red, and the yellow dotted lines represent distances in the range of 3 to 4 Å between terbinafine and surrounding residues.