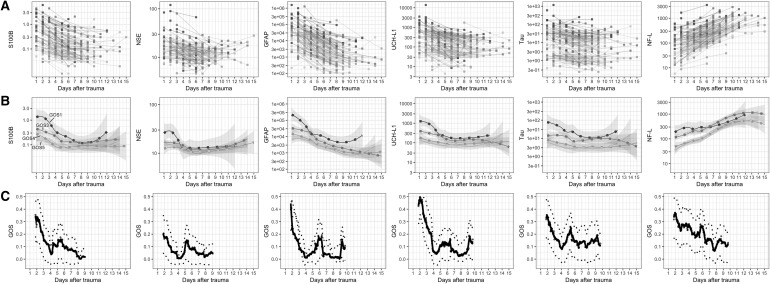
FIG. 2.
Biomarker dynamics over time stratified by outcome level. (A) Biomarker trajectories of all individuals gray graded by outcome (darker = lower GOS). The y-axis units for S100B (log, μg/L), NSE (log, μg/L), GFAP (log, pg/mL), UCH-L1 (log, pg/mL), Tau (log, pg/mL), and NF-L (log, pg/mL). (B) GOS grouped biomarker levels (group mean) over time (darker = lower GOS). The y-axis as in (A). The shown shaded 95% confidence levels are seen to widen as data become sparse. (C) Strength of association of biomarker levels toward outcome (GOS) over time (days). Univariate outcome prediction accuracy using proportional odds (Nagelkerke's pseudo-R2, y-axis) of unlogged biomarker data, within a sliding window of 200 data points is shown with a LOWESS curve fit and bootstrapped confidence interval (2 SDs). The mean of the time points in the sliding window is used, thus presenting no prediction values day 1. GFAP, glial fibrillary acidic protein; GOS, Glasgow Outcome Scale; LOWESS, LOcally WEighted Scatter-plot Smoother; NF-L, neurofilament-light; NSE, neuron-specific enolase; S100B, S100 calcium-binding protein B; SD, standard deviation; UCH-L1, ubiquitin carboxyl-terminal hydrolase-L1.