Abstract
Background
Arterial bypass and interposition grafts are used routinely across multiple surgical subspecialties. Current options include both autologous and synthetic materials; however, each graft presents specific limitations. Engineering artificial small‐diameter arteries with vascular cells derived from induced pluripotent stem cells could provide a useful therapeutic solution. Banking induced pluripotent stem cells from rare individuals who are homozygous for human leukocyte antigen alleles has been proposed as a strategy to facilitate economy of scale while reducing the potential for rejection of induced pluripotent stem cell–derived transplanted tissues. Currently, there is no standardized model to study transplantation of small‐diameter arteries in major histocompatibility complex–defined backgrounds.
Methods and Results
In this study, we developed a limb‐sparing nonhuman primate model to study arterial allotransplantation in the absence of immunosuppression. Our model was used to compare degrees of major histocompatibility complex matching between arterial grafts and recipient animals with long‐term maintenance of patency and function. Unexpectedly, we (1) found that major histocompatibility complex partial haplomatched allografts perform as well as autologous control grafts; (2) detected little long‐term immune response in even completely major histocompatibility complex mismatched allografts; and (3) observed that arterial grafts become almost completely replaced over time with recipient cells.
Conclusions
Given these findings, induced pluripotent stem cell–derived tissue‐engineered blood vessels may prove to be promising and customizable grafts for future use by cardiac, vascular, and plastic surgeons.
Keywords: animal model, arterial transplant, induced pluripotent stem cell, nonhuman primate, tissue‐engineered blood vessel, transplantation, vascular bypass
Subject Categories: Vascular Biology, Animal Models of Human Disease, Transplantation
Clinical Perspective
What Is New?
This study is the first to develop a small artery transplant model in nonhuman primates to study patency and immune response in arterial grafts, finding that (1) grafts that were partially major histocompatibility complex matched performed as well as autologous controls, (2) there was a general lack of an immune response to transplanted allografts, and (3) at the time of explant, transplanted allografts were repopulated with recipient cells.
What Are the Clinical Implications?
Cardiologists, cardiac surgeons, and vascular surgeons frequently treat patients with coronary artery disease and peripheral arterial occlusive disease, often requiring surgical intervention using small‐diameter vascular grafts, which have inherent limitations.
Someday, tissue‐engineered induced pluripotent stem cell vascular grafts may be available for clinical use; the findings of this study suggest that vascular grafts may be less immunogenic than other tissues, and if immunosuppression is required after transplantation of vascular grafts, it may only be necessary for the period of time required for the vascular graft to become populated by host cells.
Introduction
Approximately 600 000 coronary and peripheral vascular bypass procedures are performed each year in the United States.1 Arterial bypass and interposition grafts are used by multiple surgical subspecialties, including vascular surgery, cardiac surgery, neurosurgery, and reconstructive plastic surgery. Such procedures allow for redirection of blood flow around diseased arteries to reestablish circulation, reconstruct damaged blood vessels, or lengthen blood vessels for soft tissue and bony reconstruction. Current options for arterial bypass include autologous vein or arterial grafts, synthetic materials, or preserved vessels from human cadavers. Although autologous veins are the most widely used type of graft, many patients lack suitable veins for harvest, and venous grafts are prone to thrombosis, occlusion, and aneurysms caused by compliance mismatch.2 Arterial grafts often provide better long‐term patency; however, they are more challenging to harvest, are limited in length, and can be prone to vasospasm.3, 4 Moreover, harvest of venous or arterial grafts adds time, cost, and morbidity to the surgical procedure.5, 6, 7 Synthetic grafts have been successful for high‐flow, large‐diameter vessels but generally fail for smaller‐diameter vessels (<6 mm) typically used for bypass or interposition.8 Because of these barriers, there is great clinical need for readily available arterial grafts for cardiac surgery, vascular surgery, and reconstructive plastic surgery.
With induced pluripotent stem cell (iPSC) technology,9, 10 a scalable source of arterial endothelial cells and smooth muscle cells could be combined with novel scaffolds to engineer artificial small‐diameter arteries for bypass and reconstructive surgeries. Although patient‐specific iPSC‐based therapies might circumvent immune rejection, the timing and cost may preclude widespread use for many conditions. In contrast, banking of human leukocyte antigen homozygous iPSC lines could offer a strategy to provide donor‐matched arterial endothelial cells and smooth muscle cells for tissue‐engineered blood vessels.11, 12 The use of “super donors” (those individuals with type O negative blood and homozygous human leukocyte antigen alleles) may be a particularly effective approach for matching iPSC‐derived grafts to most of the population13, 14, 15 There is little known about the super donor concept with regard to primary tissue artery transplants, but even minor antigen mismatches can elicit rejection in human leukocyte antigen–matched transplants of other tissues.16, 17
Nonhuman primates provide an excellent preclinical model to study allotransplantation. Advantages to using a nonhuman primate model for preclinical studies include a long lifespan, a similar immune system to humans, and the ability to use equipment and techniques developed for human applications for cell delivery and monitoring.18, 19, 20, 21 Nonhuman primates used in biomedical research, such as rhesus and cynomolgus macaques, are usually outbred and exhibit high levels of genetic diversity. Because of this level of genetic diversity, as well as limited populations from which to select, experiments using matched tissues for transplantation studies in nonhuman primates are almost nonexistent. Fortuitously, an isolated population of cynomolgus monkeys (Macaca fascicularis) exists on the remote island of Mauritius, which has led to centuries of inbreeding and an unusually low genetic diversity. With only 7 major histocompatibility complex (MHC) haplotypes,22, 23 this Mauritian population allows for transplantation of tissues between genetically defined, matched monkeys.
Using MHC‐defined Mauritian nonhuman primates, we developed a radial artery transplant model in anticipation of studying the therapeutic value of iPSC‐derived vascular progenitors and engineered arteries. In addition to evaluating a vascular transplant model, this study examined the degree to which MHC matching may be required for future iPSC‐derived engineered blood vessel therapies and if immunosuppression would be necessary. Radial artery transplants were performed in 10 groups of monkeys with varying degrees of MHC matching (homozygous haplomatch, partial haplomatch, complete Mauritian mismatch, and Philippine/Mauritian mismatch). We assessed the patency of matched versus mismatched allografts, immunological response to allografts, and recipient versus host cell identity in allografts at the study end point. Observations from this study will help direct MHC‐histocompatibility issues related to future tissue banking and serve to demonstrate patency and durability of iPSC‐derived, small‐diameter, tissue‐engineered blood vessels.
Results
Limb‐Sparing Mauritian Cynomolgus Macaque Arterial Transplant Model
To test the patency and durability of transplanted arteries between MHC matched and mismatched donor/recipient pairs in the absence of immunosuppression, we transplanted intact radial artery segments between MHC‐defined Mauritian cynomolgus monkeys (Figure 1A through 1C). Ten pairs of animals were identified for vascular transplants, amounting to 20 allograft transplants between monkeys (40 anastomoses). These pairs included 6 homozygous haplomatches, 6 partial haplomatches, 4 complete mismatches (Mauritian cynomolgus macaques), and 4 Philippine/Mauritian complete mismatches (summarized in Table 1). Homozygous haplomatch with ABO blood type match is analogous to human super donors. A complete mismatch within Mauritian cynomolgus macaques is defined as an allograft transplant between animals with completely different haplotypes but still of Mauritian background, whereas the Philippine/Mauritian complete mismatch is an allograft transplant between one Mauritian cynomolgus macaque and one non‐Mauritian cynomolgus macaque of Philippine background; the latter represents a more divergent genetic background. We also completed 14 autograft surgical controls (28 anastomoses, summarized in Table S1).
Figure 1.
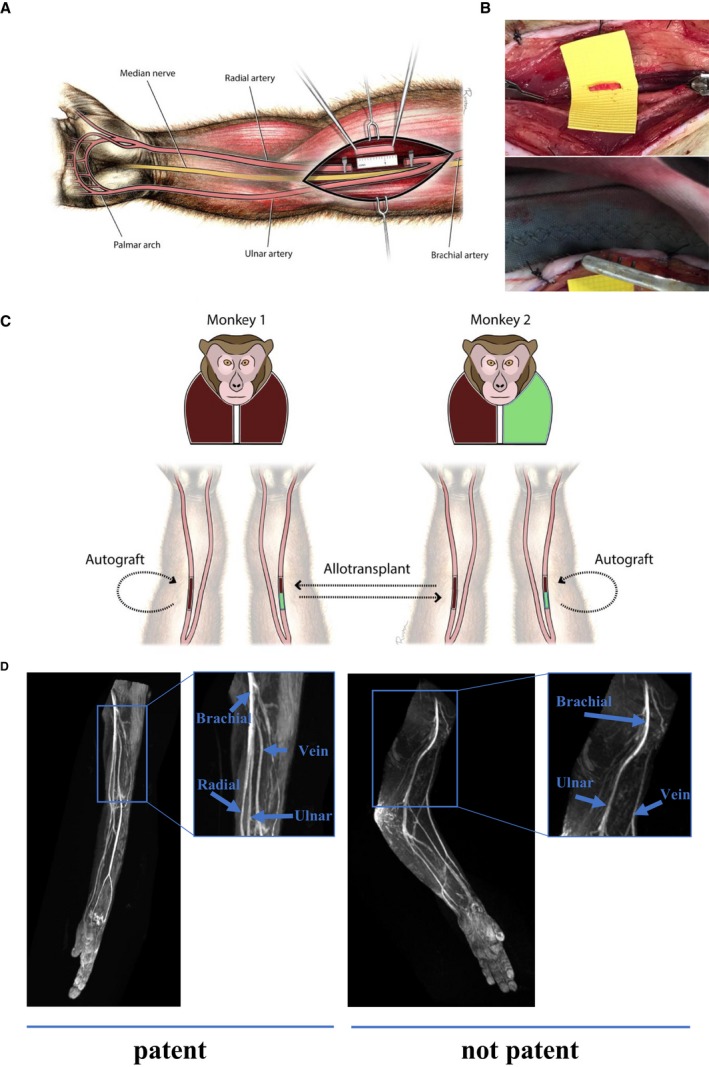
Radial artery transplant model in Mauritian cynomolgus macaques. A, Illustration demonstrating location of radial artery transplantation site in relation to the bifurcation of the brachial artery into the radial and ulnar arteries. The radial and ulnar arteries provide a redundant blood source to hand. B, Representative images of radial artery transplantation before (top) and after (bottom) microsurgical anastomosis of the radial artery graft. C, Illustration demonstrating radial artery transplant model. Monkey 1 represents a major histocompatibility complex (MHC) homozygous animal (brown/brown), and monkey 2 represents an MHC heterozygous animal (brown/green). The radial artery allograft from monkey 1 is transplanted into monkey 2 (homozygous haplomatch), whereas the radial artery allograft from monkey 2 is transplanted into monkey 1 (partial haplomatch). The autograft represents the surgical control in each monkey. D, Representative images of patent (left) and nonpatent (right) radial arteries before vessel explants. The arm with 2 patent vessels demonstrates a brachial artery that divides into the radial and ulnar arteries, which provide blood flow to the distal extremity/hand. In the arm with a nonpatent radial artery, there is no division of the brachial artery and only the ulnar artery can be traced down to hand.
Table 1.
Allograft Patency
| Animal Identifier | Sex | Haplotype | Radial Artery Swap Direction | Matching | Time After Surgery, d | Explant Patency |
|---|---|---|---|---|---|---|
| Cy0666 | Male | M1/M3 | M3/M3→M1/M3 | Homozygous haplomatch | 609 | Yes |
| Cy0675 | Female | M3/M6 | M3/M3→M3/M6 | Homozygous haplomatch | 546 | Yes |
| Cy0665 | Male | M1/M3 | M3/M3→M1/M3 | Homozygous haplomatch | 441 | Yes |
| Cy0674 | Female | M1/M3 | M3/M3→M1/M3 | Homozygous haplomatch | 490 | No |
| Cy0683 | Male | M1/M6 | M1/M1→M1/M6 | Homozygous haplomatch | 392 | No |
| Cy0681 | Male | M1/M2 | M1/M1→M1/M2 | Homozygous haplomatch | 343 | No |
| Cy0660 | Male | M3/M3 | M1/M3→M3/M3 | Partial haplomatch | 609 | Yes |
| Cy0672 | Female | M3/M3 | M3/M6→M3/M3 | Partial haplomatch | 486 | Yes |
| Cy0676 | Male | M3/M3 | M1/M3→M3/M3 | Partial haplomatch | 441 | Yes |
| Cy0661 | Female | M3/M3 | M1/M3→M3/M3 | Partial haplomatch | 462 | No |
| Cy0682 | Male | M1/M1 | M1/M6→M1/M1 | Partial haplomatch | 392 | Yes |
| Cy0680 | Male | M1/M1 | M1/M2→M1/M1 | Partial haplomatch | 343 | Yes |
| Cy0673 | Female | M1/M3 | M5/M6→M1/M3 | Mauritian mismatch | 483 | No |
| Cy0700 | Female | M5/M6 | M1/M3→M5/M6 | Mauritian mismatch | 483 | No |
| Cy0694 | Female | M3/M3 | M4/M4→M3/M3 | Mauritian mismatch | 364 | Yes |
| Cy0695 | Female | M4/M4 | M3/M3→M4/M4 | Mauritian mismatch | 364 | No |
| Cy0692 | Female | M2/M4 | NMC→M2/M4 | PHL/MAU mismatch | 259 | No |
| Cy0613 | Female | NMC | M2/M4→NMC | PHL/MAU mismatch | 259 | No |
| Cy0667 | Female | M3/M4 | NMC→M3/M4 | PHL/MAU mismatch | 308 | No |
Mauritian mismatch indicates complete mismatch within Mauritian background. NMC indicates non‐Mauritian cynomolgus; PHL/MAU mismatch indicates Philippine/Mauritian mismatch.
Homozygous Haplomatched/Partial Haplomatched Allografts Demonstrate Similar Patency to Surgical Controls
Before graft explant, magnetic resonance angiograms were performed to determine graft patency (Figure 1D). When the grafts were explanted, at least 6 months after autotransplantation or allotransplantation, there was no difference in patency between homozygous haplomatch (50%) and partial haplomatch (83%) grafts (P=0.27) (Tables 1 and 2). Furthermore, surgical control grafts (67%) had similar patency (Table 2 and Table S1) to both homozygous haplomatch (P=0.62) and partial haplomatch (P=0.26). Completely mismatched grafts (including both Mauritian and Philippine mismatch groups) had the lowest patency rates (14%); however, they were not statistically significant compared to homozygous haplomatch (P=0.26) and surgical controls (P=0.15) (Tables 1 and 2). There was a significant difference in patency between partially haplomatched grafts and complete mismatched grafts (P=0.03) (Table 2). These data suggest that homozygous/partially matched and autologous matched transplants are equivalent. Of note, 1 of 7 completely mismatched grafts remained patent at 1 year, and 8 of 12 haplomatched grafts remained patent without immunosuppression for a year or more (Tables 1 and 2).
Table 2.
Allograft/Autograft Summary
| Graft | Patency, No./Total (%) | Lumen Area, median (IQR), mm2 a , b | Intima/Media Ratio, median (IQR)a , b |
|---|---|---|---|
| Allograft homozygous haplomatch | 3/6 (50) | 0.52 (0.24) | 4.3 (4.3) |
| Allograft partial haplomatch | 5/6 (83)c | 0.17 (0.42) | 2.4 (4.0) |
| Allograft complete mismatch | 1/7 (14)c | 0.15 (NA) | 8.1 (NA) |
| Autograft surgical control | 6/11 (55) | 0.14 (0.15) | 1.7 (1.5) |
IQR indicates interquartile range; NA, not available because of only one patent vessel (one measurement).
Only measured in patent grafts.
No statistical significance between any of the groups.
P=0.03, 2‐sided Fisher's exact test.
Patent vessels showed variable degrees of intimal hyperplasia, extracellular matrix deposition, and elastic lamina fragmentation and multiplication (Figure 2A). These changes were observed across all groups, with no noticeable differences between matched and mismatched grafts. Morphologic changes in nonpatent vessels were more variable, including recanalized thrombus or elastic lamina without typical arterial layers indicative of atrophic changes (Figure 2A). Infiltration of the grafts with lymphoid cells, macrophages, or granulocytes was not observed. Similar morphological changes were observed in the control autografts and across experimental groups. In 10 of 15 graft failures (67%), including autograft failures, a pattern of a patent proximal anastomosis, followed by a thin fibrotic graft, was noted (Figure 2B).
Figure 2.
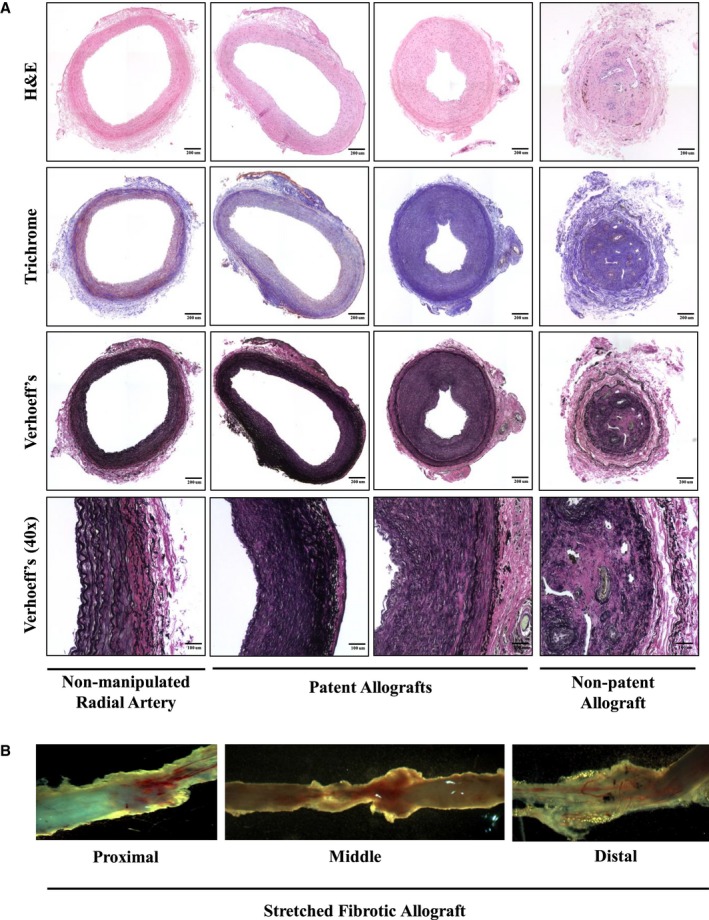
Pathological analysis of allografts. At the end of the study, radial artery grafts were explanted. The proximal anastomosis was fixed and stained. The distal anastomosis was frozen in O.C.T. A, Representative images of radial artery allografts from a normal nonmanipulated, patent allograft, and nonpatent vessels, respectively. Hematoxylin and eosin (H&E), Trichrome (fibrosis), and Verhoeff's (elastin) stains show pathological changes observed in allografts compared with normal artery. These include near normal (left patent allograft), intimal hyperplasia (right patent allograft), fibrosis (patent and nonpatent allograft), elastic lamina multiplication (patent and nonpatent allograft), and recanalized thrombus obstructing the artery lumen (nonpatent allograft). First three panels: ×20 magnification scanned images (bar=200 μm). Last panel: ×40 magnification scanned image (bar=100 μm). B, Representative images of fibrotic stretched out allograft from proximal to distal end.
Lack of Detectable Immune Response to Matched and Mismatched Grafts
To test for persistent immunologic memory, which might allow for discrimination between graft failure caused by transplant rejection versus surgical failure, the presence of donor‐specific antibody (DSA) after transplantation was tested. Only 1 of 19 monkeys developed detectable DSA. Cy0700 (M5/M6) generated antibodies to the Cy0673 (M1/M3) allograft (Figure 3A and Table S2 and Figure S1). Interestingly, Cy0673 did not develop antibodies to Cy0700, despite there being an MHC mismatch in that direction. A mixed lymphocyte reaction was performed to determine whether there was any evidence of cellular‐mediated immune responses in the transplant pairs.24 Although varying degrees of T‐cell proliferation between the haplomismatched pairs were detected (Table 3), there was no correlation between the extent of proliferation and long‐term graft patency (Figure 3B and Table 1). In addition, there was no association between the magnitude of the mixed lymphocyte reaction proliferative response and the development of DSA (Figure 3A and 3B).
Figure 3.
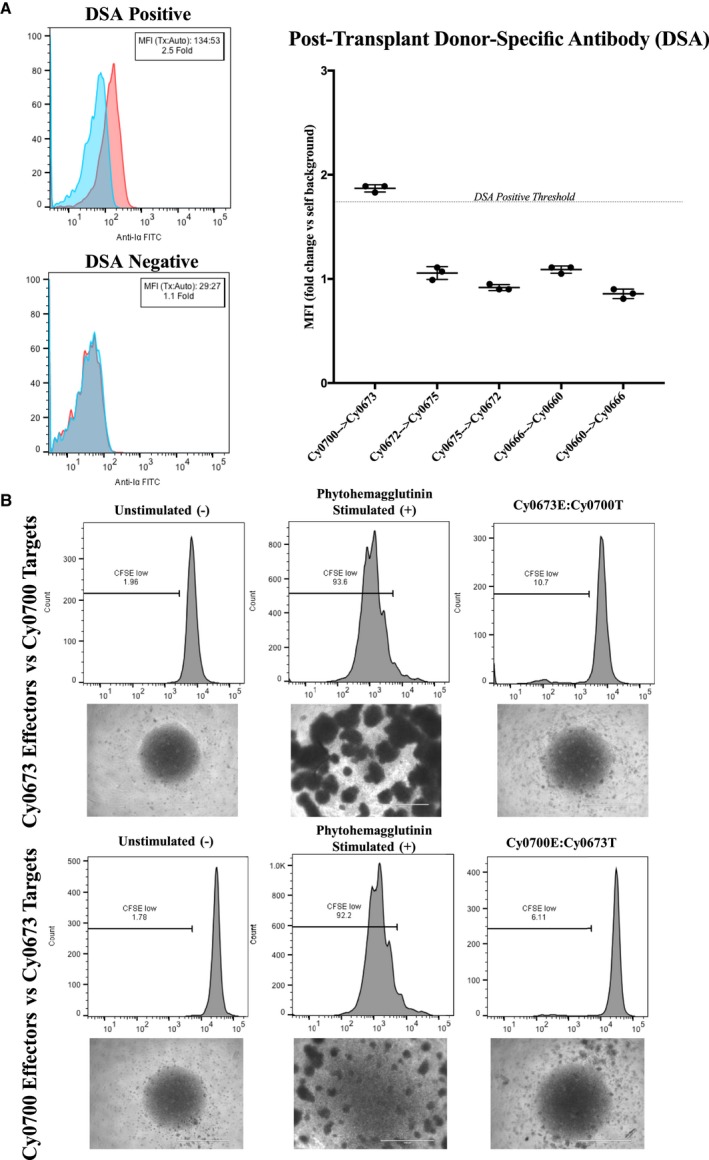
Assessment of de novo donor‐specific antibody (DSA) development and mixed lymphocyte reactions. Blood serum samples were drawn from animals pretransplant and posttransplant to assess for the presence of preexisting and de novo DSAs, respectively. Serum was incubated with autologous or transplant donor peripheral blood mononuclear cells (PBMCs), washed, and then stained with secondary antimacaque IgG–fluorescein isothiocyanate antibody to detect bound DSA (n=3 replicates per sample). A, Left, Representative flow cytometric staining of a DSA‐positive transplant recipient (Cy0700) shows a shift in median fluorescence intensity (MFI) >1.75× over autologous background. Data are representative of n=5 repeated experiments. Right, Representative graph summarizing the results of one experiment, including DSA‐positive and DSA‐negative animals. B, One‐way mixed lymphocyte reactions. Top, Target PBMCs from transplant donor Cy0700 were irradiated (30 Gy) and labeled with CellTrace‐Violet dye, whereas effector PBMCs from transplant recipient Cy0673 were not irradiated and labeled with CFSE (carboxyfluorescein succinimidyl ester) proliferation dye. Images of culture wells were taken showing the unstimulated effector PBMCs, those effectors stimulated with T‐cell mitogen phytohemagglutinin, and the cocultured effectors/targets. Bottom, Target and effector were switched for the same transplant pair and analyzed as above. Data are representative of triplicate wells per condition.
Table 3.
Mixed Lymphocyte Reaction
| Animal Identifier | Sex | Haplotype | Assay Direction | Mean CFSElo, % | SD |
|---|---|---|---|---|---|
| Cy0660 | Male | M3/M3 | Cy0666E→Cy0660T | Assay fail | Assay fail |
| Cy0666 | Male | M1/M3 | Cy0660E→Cy0666T | 1.42 | 0.15 |
| Cy0661 | Female | M3/M3 | Cy0674E→Cy0661T | 1.51 | 0.02 |
| Cy0674 | Female | M1/M3 | Cy0661E→Cy0674T | 1.32 | 0.07 |
| Cy0673 | Female | M1/M3 | Cy0700E→Cy0673T | 6.17 | 0.18 |
| Cy0700 | Female | M5/M6 | Cy0673E→Cy0700T | 11.03 | 1.72 |
| Cy0694 | Female | M3/M3 | Cy0695E→Cy0694T | 14.13 | 1.92 |
| Cy0695 | Female | M4/M4 | Cy0694E→Cy0695T | 16.53 | 1.96 |
| Cy0692 | Female | M2/M4 | Cy0613E→Cy0692T | 4.44 | 0.08 |
| Cy0613 | Female | NMC | Cy0692E→Cy0613T | 6.81 | 1.95 |
A summary of transplant effector/target pair mixed lymphocyte reaction detection is shown. Assay direction indicates effector (E) peripheral blood mononuclear cell (PBMC) proliferation to target (T); CFSE (Carboxyfluorescein succinimidyl ester); PBMC (effector PBMC→target PBMC). Animals are grouped by transplant pairs. NMC indicates non‐Mauritian cynomolgus.; CFSELo, Carboxyfluorescein succinimidyl ester low expression.
Allografts Become Remodeled Over Time
Because most arterial allografts remained in place for over 1 year, it was important to determine whether donor vascular cells were still present at the time of explant. Cells from the middle of each graft were dissociated and then sorted by fluorescence‐activated cell sorting (FACS) for the presence or absence of the endothelial marker CD31 (platelet endothelial cell adhesion molecule 1) (Figure S2). Genomic DNA (gDNA) was isolated from each sorted cell population and analyzed for single‐nucleotide polymorphisms (SNPs) unique to each animal to determine the cell's identity (Figure S2). In all cases, cells taken from the autograft surgical controls were recipient cells (Figure S2). In allografts, CD31‐positive cells taken from the donor grafts contained all recipient SNPs or a mixture of recipient and donor SNPs (Figure 4). More surprisingly, in all cases, the CD31‐negative population contained all recipient SNPs (Figure 4). These data demonstrate that (1) donor grafts become remodeled and repopulated by recipient cells and (2) endothelial cells sometimes have the ability to persist for over a year in allograft vessels.
Figure 4.
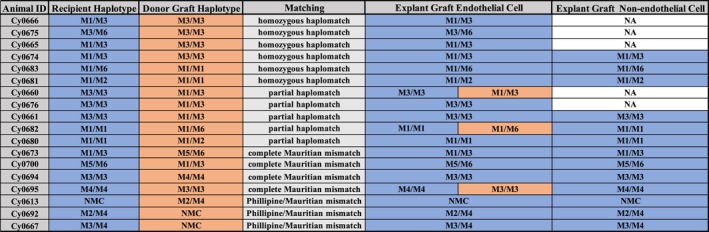
Summary table of single‐nucleotide polymorphism analysis of host/donor cells in allografts. Orange represents donor graft haplotype. Blue represents recipient haplotype. NA, the CD31‐negative cells were not sorted in those samples.
Discussion
We developed a small‐artery transplant model in Mauritian cynomolgus macaques to test the long‐term function of transplanted arteries in MHC matched and mismatched animals without immunosuppression. This model provides a reproducible small‐artery transplant technique, easy postoperative graft monitoring, and the ability of the grafts to fail and/or be explanted for immunologic studies without compromising extremity blood flow and subjecting the animals to significant morbidity. The results of this study show that partially haplomatched arterial grafts performed as well as autologous surgical controls. In addition, there was a lack of detectable immune response to arterial allografts, and over the course of the study, transplanted allografts became remodeled with recipient cells.
Given the final outcomes of the grafts, it appears that partial haplomatching (ie, ensuring that donor and recipient share a common haplotype) may be enough to yield a successful outcome. Autologous surgical controls, homozygous haplomatched grafts, and partially haplomatched grafts show similar patency rates to what has been previously reported in human peripheral bypass studies with radial arteries. For example, radial artery grafts used for infrainguinal bypass had 62% patency after 1 year and 56% patency at 3 to 5 years, whereas below‐the‐ankle bypass with radial artery yielded 83% patency at 3 years.25, 26 The present study demonstrates a similar finding: even with a perfectly matched radial artery (autologous surgical control) we still observed only 67% patency for a minimum of 1 year after anastomosis.
The pathological characteristics between failed allografts and autografts were similar, and immune infiltration was not detected in histological samples of explants. The shortest time between transplant and explant was 259 days. It is possible that by this time an acute immune response could have destroyed the graft with no evidence of persisting chronic rejection. Indeed, the positive mixed lymphocyte reaction result for the DSA‐positive animal (Cy0700) may indicate that an acute rejection response contributed to graft failure in this animal. However, for the other transplant pairs tested, we found no evidence of rejection by DSA assay, mixed lymphocyte reaction assay, or histological analysis. Given our observations of autologous graft failure in the surgical controls, it is more likely that the graft failures observed in this model are potentially the result of the surgical procedure itself. One limitation to this model is that microsurgical anastomoses are technically challenging, particularly those <2 mm. In addition, other surgical factors that could potentially contribute to graft failure include a high observed resting tension and elasticity of the radial artery in nonhuman primates, possibly attributable to evolutionary demands and the need for significant elastic recoil in neurovascular structures in the upper extremity; given this, the anastomoses could potentially be burdened by the high tension at rest, leading to fibrosis and thinning. Our lack of observed immune responses in this nonimmunosuppressed environment may indicate that these vascular grafts display a lower degree of immunogenicity compared with other tissue types (Figure 3A and 3B),27, 28 but we have not yet examined the early in vivo immune response to allogenic and autologous vascular cells.29
To explore a donor or recipient identity of smooth muscle and endothelial cells in the radial artery allografts at the time of graft explant, we analyzed gDNA from CD31‐positive endothelial cells and CD31‐negative cells (most of which are likely smooth muscle cells) from each of the grafts. In all allografts, the donor CD31‐negative cells were replaced with recipient CD31‐negative cells. In all but 3 cases, donor CD31‐positive cells were replaced with recipient CD31‐positive cells. The remaining 3 grafts exhibited a heterogeneous population of recipient and donor CD31‐positive cells. Thus, the graft tissue is mostly replaced with recipient cells, which could originate from either cells in the vascular tissue flanking the graft or vascular progenitor cells in circulation.30, 31, 32, 33, 34, 35
The replacement of donor cells with recipient cells has previously been demonstrated in transplant arteriosclerosis.36, 37 Transplant arteriosclerosis is characterized by vascular lesions in the graft consisting of concentric myointimal proliferation. Previous studies examining the origin of vascular smooth muscle cells in transplant arteriosclerosis found donor as well as recipient origins of the graft vessel endothelium, and recipient cells contributed significantly to neointimal hyperplasia. In contrast, rodent models of vascular transplantation have demonstrated high percentages of recipient cells after transplantation, but these studies typically do not include immunosuppression.30, 36, 38, 39, 40, 41, 42 Although these studies examined replacement of smooth muscle cells, replacement of endothelial cells has been more difficult to study in vivo. Recently, a possible mechanism by which endothelial cells are replaced after injury has been described.42
One potential limitation to this model is the clinical relevance to vascular disease requiring vascular bypass because both donor and recipient subjects were generally healthy, without a diganosis of peripheral arterial occlusive disease. However, we believe that our data are informative in helping understand fundamental aspects of primary blood vessels in a transplant context. More specifically, the primary focus of these studies was to understand the degree of MHC matching that would be required in future studies using iPSCs. There is little information in the literature on the immune response to transplanted vascular grafts in the clinical setting. Vascular bypass surgeries are done using autologous artery or vein in most cases or synthetic material with or without autologous vascular cells from the patient. Given limitations in autologous adult cells, various stem cell sources have been explored for vascular tissue engineering.43 Using allogenic iPSCs could eliminate the problems with cell quality and variations associated with patient‐specific therapies and remove delays in graft availability, potentially making grafts off‐the‐shelf. There is a body of literature describing primary grafts and tissue‐engineered grafts in animal models.43 More specifically, remodeling of transplanted tissue‐engineered vascular grafts with host endothelial cells has been described, similar to what we describe with primary vascular grafts.44 Clinically, the immune response to allogenic tissue has not been explored in depth; however, the lack of an immune response has been reported previously, similar to the findings in the present study.45, 46, 47, 48 We hypothesize that the lack of immune response that we observed in all but one of our animals was caused by the remodeling of the graft with host cells.
Transplanting native radial arteries between matched and partially matched animals resulted in similar patency rates as the autologous controls at least 1 year after transplantation. Surprisingly, transplanting completely mismatched radial arteries resulted in only one animal developing a DSA response. Because most of our allografts were mostly comprised of recipient cells at the time of explant, it is possible that this turnover mitigated any immune response to alloantigens. The data presented in this study suggest that the use of iPSC bioengineered arterial constructs for therapy may only require the attenuation of the immune response until the graft becomes repopulated by recipient cells.
Materials and Methods
The authors declare that all supporting data are available within the article (and its online supplementary files).
Animal Model
All Mauritian cynomolgus macaques were housed at the Wisconsin National Primate Research Center. All procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and under the approval of the University of Wisconsin–Madison College of Letters and Sciences and the Vice Chancellor Office for Research and Graduate Education Centers (LSVC [Letters and Sciences Vice Chancellor]) Animal Care and Use Committee. A total of 20 Mauritian cynomolgus macaques were used in this study, composed of pairs for homozygous haplomatch and partial haplomatch comparison (12 animals), Mauritian mismatch comparison (4 animals), and Philippine/Mauritian mismatch comparison (4 animals).
Statistical Analysis
A 2‐sided Fisher's exact test was used to determine the significance in patency rates between each group (ie, homozygous haplomatch versus partial haplomatch). A 2‐sided Wilcoxon rank‐sum test was used to determine significance of lumen area and intima/media ratio between each group (ie, homozygous haplomatch versus partial haplomatch). Only patent vessels were used to determine statistical significance of lumen area and intima/media ratio. The P‐value cutoff for statistical significance used was 0.05. Statistical analysis was performed using the Mstat program (https://mcardle.wisc.edu/mstat/).
MHC Typing
Genotyping of MHC‐I and MHC‐II genes in M fascicularis (cynomolgus monkeys) was performed as previously described.22 Briefly, gDNA was isolated from peripheral blood mononuclear cell samples and used as templates for polymerase chain reaction with a panel of primers that flank the highly polymorphic peptide binding domains encoded by exon 2 of class I (Mafa‐A, Mafa‐B, Mafa‐I, and Mafa‐E) and class II (Mafa‐DRB, Mafa‐DQA, Mafa‐DQB, Mafa‐DPA, and Mafa‐DPB) loci. These polymerase chain reaction products were generated with a Fluidigm Access Array, which allows all reactions to be multiplexed in a single experiment. After cleanup with AMPure beads and pooling, the amplicons were sequenced on an Illumina MiSeq instrument; and the resulting sequence reads were mapped against custom databases of Mauritian cynomolgus macaque class I and class II sequences. Because each sequence read is tagged with a unique set of barcodes, it can be traced back to the animal from which it originated. This approach provides comprehensive genotypes for both class I as well as the DRB, DQA/DQB, and DPA/DPB class II loci.
Imaging
Before graft explant, animals underwent magnetic resonance angiography imaging to identify graft patency before harvest and assess distal blood flow before explant. Animals were anesthetized, transferred to an imaging facility, and placed in a standard immobilization position. Scans were performed on the GE Signa positron emission tomographic/magnetic resonance instrument (3 T). Multihance contrast was loaded at 0.2 mmol/kg and diluted to 10 mL. Injections of 5 mL multihance contrast, followed by a 20‐mL flush injected at 1.5 mL/s each, were performed for vascular imaging. During tricks scan of 15 phases (25 seconds into the scan), we injected at the pause after the first phase. Precontrast and postcontrast FSPGR (fast spoiled gradient echo) subtraction was performed. Images were viewed and analyzed using Horos software (horosproject.org).
Surgical Technique
All surgeries were performed in accordance with Institutional Animal Care and Use Committee guidelines under the supervision of trained veterinary staff. Surgeries were performed with animals under general anesthesia with an endotracheal tube. All animals were positioned supine with the operative extremities(s) secured at ≈30° of shoulder abduction and 60° of elbow flexion. Positioning the animal in this manner minimized tension on the arterial anastomoses while simultaneously providing exposure of the vasculature of the upper arm. Optimal extremity position was determined via anatomic cadaver dissections with 2 animals not included in this study.
Arterial transplantation
For both radial artery allotransplants and autotransplants, a 6‐ to 8‐cm incision was made over the medial aspect of the arm, along the interval between the biceps and triceps muscles. Subcutaneous flaps were elevated, and the intermuscular septum was dissected. The brachial, radial, and ulnar arteries were identified. In the M fascicularis species, the brachial bifurcation was found at the mid‐upper arm in all subjects. The brachial, radial, and ulnar arteries were separated from the adjacent vena comitans. Using standard microvascular techniques, microvascular clamps were applied to the brachial artery and radial artery distal to the planned graft resection. An additional clamp was applied to the proximal ulnar artery to prevent retrograde flow into the operative field. On average, a 13‐mm segment of radial artery starting 3 mm distal to the brachial‐radial artery bifurcation was then sharply excised. The excised segment was removed from the operative field and kept sterile in saline solution at room temperature for no more than 5 minutes.
The excised arterial segments were intended to be immediately transplanted (ie, no more than 5 minutes in room temperature sterile saline solution) as interposition radial artery grafts. These were either explanted and reimplanted from the same arm (autotransplant/autograft control) or explanted from one monkey and implanted into another monkey (allotransplant/allograft). Before performing the anastomoses, each animal was administered 200 units of intravenous heparin. Using the operating microscope, the proximal and distal anastomoses were sewn using simple interrupted 9‐0 Nylon sutures (Ethicon Inc, Somerville, NJ). Topical heparinized saline (concentration, 100 U/mL) was used during vessel preparation and suturing of the anastomose. All anastomoses were confirmed patent using a vascular strip test. Additional sutures and anastomotic revisions were performed as needed to achieve anterograde flow through the radial artery graft. Hemostasis was achieved using electrocautery, and the incision was closed in 2 layers using polyglactin absorbable sutures. The animals were awoken from anesthesia and extubated.
Harvest of transplanted segments
For histopathologic assessment, we selected an end point of at least 6 months after the initial transplant. This end point was selected to allow for sufficient time to observe subacute or chronic inflammation, fibrosis, and/or signs of graft rejection; and it is comparable to end points of other studies of vascular graft patency.49, 50 In several cases, the grafts remained in place for ≈1 year on the basis of surveillance imaging findings, suggesting that the grafts were patent (data not shown). Explantation was performed under general anesthesia. The previous incision was incised, and the radial artery was identified. Careful inspection under the operating microscope allowed for identification of the proximal and distal anastomoses. Inspection, strip test, and sterile Doppler probe were used to assess arterial flow through the previous anastomoses. The artery to be excised was then clamped, and distal perfusion was assessed. The radial artery was ligated proximal and distal to the transplanted segment, and the segment was sharply excised. The incision was closed using 2 layers of absorbable sutures, and the animal was awoken from anesthesia.
Arterial Graft Postexplant Processing
Adventitia was cleaned from the graft, and lumen was flushed of any remaining blood. A 2‐ to 3‐mm section, starting at the visible suture site and extending 2 to 3 mm into the donor graft of the proximal and distal anastomosis sites, was taken. The proximal anastomosis site up to 3 mm length was dissected and fixed in 4% paraformaldehyde, paraffin embedded, and sectioned for histopathological analysis. The distal anastomosis site up to 3 mm length was dissected and embedded in Tissue Tek OCT compound (Sakura catalog No. 4583) for frozen sections in case of need. The middle of the graft was analyzed for donor versus recipient cell identity. Briefly, the tissue was digested in collagenase/dispase for 1 to 2 hours until most of the tissue was dissociated. Dissociated cells were washed in 2% fetal bovine serum in PBS and stained for CD31 (clone WM59 BD Biosciences catalog No. 555446). CD31‐positive and CD31‐negative populations were analyzed and sorted on the BD FACSARIA II. DNA was isolated from sorted cell populations for SNP chimerism assay using the Zymo Quick‐DNA plus kit (Zymo Research, D4074).
Histopathological Analysis
The proximal anastomosis sites were embedded in paraffin wax and cut into cross‐sections of 10 μm in sequential order and mounted onto microscopic slides. Proximal radial artery sections were stained using hematoxylin and eosin, Trichrome for fibrosis, and Verhoeff's stain for elastin. All stains were performed by the TRIP pathology laboratory (University of Wisconsin–Madison School of Medicine and Public Health, Department of Pathology and Laboratory Medicine). Histological analyses of hematoxylin and eosin, Trichrome, and Verhoeff's stains were performed by an independent pathologist (I.I.S.) at the Wisconsin National Primate Research Center. Hematoxylin and eosin sections were used to examine lumen area and intima/media ratio with ImageJ software. Lumen area is defined as the interior area of patent vessels (expressed in mm2). We defined the intima/media ratio as a ratio of the measurements of the thickness of the tunica intima/tunica media. The lumen area and intima/media ratio were analyzed only in patent vessels for statistical analysis.
SNP Chimerism Assay
SNP selection
Twelve SNPs were selected from previously published data identifying SNPs and allele frequency in ≈250 Mauritian cynomolgus macaques.51 Selection criteria for SNPs included the following: (1) minor allele frequencies between 0.35 and 0.55; (2) SNPs were at least 42 Mb apart (11/12 on separate chromosomes); (3) all SNPs were classified as synonymous mutations; and (4) availability of published M fascicularis sequence for gene containing SNP. A similar previously published human assay found that if SNPs had a minimal allele frequency of 0.4, then there was a 99.6% likelihood of at least one SNP being different between 2 animals when comparing 11 SNPs.52
Graft sample SNP identification
A titration was done with 1 ng, 100 pg, 50 pg, 30 pg, 20 pg, 10 pg, and 5 pg of gDNA per reaction. A water‐only negative control was also included. Once informative SNPs were identified in donor/recipient pairs, all samples isolated from grafts were run in triplicate, according to manufacturer's instructions, with the exception of using 30 pg of gDNA per reaction and performing the assay with 46 rather than 40 cycles. For all samples, gDNA isolated from blood and previously used for SNP genotyping was diluted to 30 pg per reactions for both donor and recipient animals used as controls. In addition, a mixture of donor and recipient gDNA was combined, the sample was diluted to 30 pg per reaction for use as a heterozygous control, and Millipore water was used as a negative control. Data were analyzed using “end point genotyping” data analysis Roche LightCycler 96 Instrument software.
SNP genotyping
gDNA was isolated from whole blood (with EDTA) using the Qiagen DNeasy Blood & Tissue Kit (catalog No. 69504), as per manufacturer's protocol. After isolation, gDNA concentration was determined using ThermoFischer Qubit dsDNA HS Assay (catalog No. Q32851). The IDT rhAmp SNP Assay was used to SNP genotype all animals. Custom primers were designed for all 11 SNPs using IDT's online software. All genotyping reactions were run in triplicate, as per manufacturer's instructions, using a final concentration of 2 ng gDNA per reaction. The Roche LightCycler 96 Instrument was used to record VIC and FAM intensity after every polymerase chain reaction cycle. The end point genotyping data analysis option on instrument software was used to identify SNP genotype on the basis of end point fluorescence of VIC and FAM. The SNP genotype for donor/recipient pairs was compared to identify informative SNPs. Informative SNPs refers to SNPs that had different genotypes in the donor and recipient animals; when possible, SNPs that were homozygous for different SNPs in donor/recipient pairs were used.
DSA Assay
Blood serum samples were drawn using BD SST tubes (BD Biosciences, Franklin Lakes, NJ) from animals pretransplant and posttransplant to assay for presence of preexisting and de novo DSAs, respectively. Serum samples were frozen at collection day and thawed fresh on the day of assay. Peripheral blood mononuclear cells were collected in CPT tubes (BD Biosciences), processed according to manufacturer's instructions, and cryopreserved in either fetal bovine serum + 10% dimethyl sulfoxide or CryoStor CS10 freezing medium (Stem Cell Technologies, Vancouver, CA) until thawed on day of the assay. Serum was diluted 1:5 in FACS Buffer, incubated for 30 to 90 minutes with 2 × 105 autologous or transplant donor peripheral blood mononuclear cells + FcR block (BD Biosciences), washed twice, and then stained with secondary antimacaque IgG–fluorescein isothiocyanate antibody (clone 1B3; Nonhuman Primate Reagent Resource, Worcester, MA) to detect bound DSA. Dead cells were excluded with BD Horizon Fixable Viability Stain 700, then stained and gated for T cells (ie, CD3+ [PerCPCy5.5, clone SP34‐2] and CD20‐ [APC, clone 2H7]) for DSA analysis. Samples were acquired on a BD FACSCANTO II flow cytometer and analyzed using FlowJo software v10 (BD Biosciences). Each experiment used triplicate samples for each condition. DSA‐positive samples were repeated and reproduced in 5 of 5 separate experiments to ensure validity. Because of the low SD between intraexperimental sample replicates, a threshold for DSA positivity of >1.75‐fold increase in fluorescein isothiocyanate median fluorescence intensity compared with autologous background was chosen on the basis of a positive control rhesus sample (data not shown). Because of fluctuations in flow cytometer settings and performance, sample comparisons were only conducted within individual experiments versus DSA‐positive control.
Mixed Lymphocyte Reaction
Target peripheral blood mononuclear cells from transplant donor were irradiated (30 Gy) and labeled with CellTrace‐Violet dye (ThermoFisher catalog No. 562158), whereas effector peripheral blood mononuclear cells from transplant recipient were not irradiated and labeled with CFSE (carboxyfluorescein succinimidyl ester) proliferation dye (ThermoFisher catalog No. 565082). Cells were cocultured at a 1:1 ratio for 5 days in 96‐well tissue culture plates in ImmunoCult‐XF serum‐free T‐cell expansion medium (StemCell Tech catalog No. 10981) with 100 IU/mL rhIL2 (recombinant human interleukin 2). On the day of harvest, cells were collected and stained with BD Horizon Fixable Viability Stain (BD Biosciences catalog No. 564407) and antiprimate CD3 (BD Biosciences catalog No. 557597). Viable singlet CD3+Violet‐ cells were analyzed for CFSE dilution.
Sources of Funding
Research reported in this publication was supported in part by the Office of The Director, National Institutes of Health (NIH) under Award P51OD011106 to the Wisconsin National Primate Research Center, University of Wisconsin–Madison; in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award T32AI125231; and in part by NIH/National Heart, Lung, and Blood Institute grant U01HL134655. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Disclosures
None.
Supporting information
Table S1. Autograft Patency
Table S2. Donor Specific Antibody (DSA)
Figure S1. Pre‐transplant donor‐specific antibodies. Representative graph summarizing the pre‐transplant results of one experiment showing DSA positive pair Cy0700 and Cy0673.
Figure S2. Assessment of cell identity within allografts. At end of study, radial artery grafts were explanted. The proximal and distal anastomosis sites were saved for sectioning and staining while the middle of the graft was processed and analyzed for the presence of host/donor cells. A, The graft was digested and individual graft cells were sorted for CD31+ and CD31‐ populations. Shown is a representative sort for CD31 cells. B, Plot of SNP analysis results from all samples in the study. C, Plot of SNP analysis results from a partial haplomatch (left panel) and complete mismatch animal (right panel) that showed a mixture host and donor endothelial cells within the graft.
Acknowledgments
Special thanks to Wisconsin National Primate Research Center services, including scientific protocol implementation staff, surgical technicians, veterinary services, immunology services, and the genetic services unit for providing major histocompatibility complex genotyping. Special thanks to the University of Wisconsin radiology and imaging facility, specifically, Sarah Kohn (ultrasound) and Kelly Hellebrant (magnetic resonance imaging). Thanks to the Morgridge Institute for Research. Thanks to Jue Zhang for assisting in experiments and to David Vereide and William Burlingham for reviewing the manuscript. Author Contributions J.P.M., M.E.B., L.E.K., and E.S.P. performed experiments. J.P.M., M.E.B., and L.E.K. analyzed results and made figures. S.J.K., J.S.I., N.J.A., W.Z., and S.O.P. performed surgical procedures and assisted with manuscript preparation. R.J.S. created scientific illustrations and assisted with manuscript preparation. I.I.S. performed pathology diagnosis. J.P.M., J.A.T., and S.O.P. designed research. J.P.M., J.A.T., and S.O.P. wrote the manuscript.
(J Am Heart Assoc. 2019;8:e012135 DOI: 10.1161/JAHA.119.012135.)
References
- 1. Ravi S, Qu Z, Chaikof EL. Polymeric materials for tissue engineering of arterial substitutes. Vascular. 2009;17(suppl 1):S45–S54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Campbell GR, Campbell JH. Development of tissue engineered vascular grafts. Curr Pharm Biotechnol. 2007;8:43–50. [DOI] [PubMed] [Google Scholar]
- 3. Acar C, Jebara VA, Portoghese M, Beyssen B, Pagny JY, Grare P, Chachques JC, Fabiani JN, Deloche A, Guermonprez JL. Revival of the radial artery for coronary artery bypass grafting. Ann Thorac Surg. 1992;54:652–659; discussion 659–660. [DOI] [PubMed] [Google Scholar]
- 4. Carpentier A, Guermonprez JL, Deloche A, Frechette C, DuBost C. The aorta‐to‐coronary radial artery bypass graft: a technique avoiding pathological changes in grafts. Ann Thorac Surg. 1973;16:111–121. [DOI] [PubMed] [Google Scholar]
- 5. Verma S, Szmitko PE, Weisel RD, Bonneau D, Latter D, Errett L, LeClerc Y, Fremes SE. Should radial arteries be used routinely for coronary artery bypass grafting? Circulation. 2004;110:e40–e46. [DOI] [PubMed] [Google Scholar]
- 6. McKee JA, Banik SS, Boyer MJ, Hamad NM, Lawson JH, Niklason LE, Counter CM. Human arteries engineered in vitro. EMBO Rep. 2003;4:633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berger PB, Alderman EL, Nadel A, Schaff HV. Frequency of early occlusion and stenosis in a left internal mammary artery to left anterior descending artery bypass graft after surgery through a median sternotomy on conventional bypass: benchmark for minimally invasive direct coronary artery bypass. Circulation. 1999;100:2353–2358. [DOI] [PubMed] [Google Scholar]
- 8. Catto V, Fare S, Cattaneo I, Figliuzzi M, Alessandrino A, Freddi G, Remuzzi A, Tanzi MC. Small diameter electrospun silk fibroin vascular grafts: mechanical properties, in vitro biodegradability, and in vivo biocompatibility. Mater Sci Eng C Mater Biol Appl. 2015;54:101–111. [DOI] [PubMed] [Google Scholar]
- 9. Yu J, Vodyanik MA, Smuga‐Otto K, Antosiewicz‐Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. [DOI] [PubMed] [Google Scholar]
- 10. Yu J, Hu K, Smuga‐Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beatty PG, Boucher KM, Mori M, Milford EL. Probability of finding HLA‐mismatched related or unrelated marrow or cord blood donors. Hum Immunol. 2000;61:834–840. [DOI] [PubMed] [Google Scholar]
- 12. Faden RR, Dawson L, Bateman‐House AS, Agnew DM, Bok H, Brock DW, Chakravarti A, Gao XJ, Greene M, Hansen JA, King PA, O'Brien SJ, Sachs DH, Schill KE, Siegel A, Solter D, Suter SM, Verfaillie CM, Walters LB, Gearhart JD. Public stem cell banks: considerations of justice in stem cell research and therapy. Hastings Cent Rep. 2003;33:13–27. [PubMed] [Google Scholar]
- 13. Nakatsuji N, Nakajima F, Tokunaga K. HLA‐haplotype banking and iPS cells. Nat Biotechnol. 2008;26:739–740. [DOI] [PubMed] [Google Scholar]
- 14. Turner M, Leslie S, Martin NG, Peschanski M, Rao M, Taylor CJ, Trounson A, Turner D, Yamanaka S, Wilmut I. Toward the development of a global induced pluripotent stem cell library. Cell Stem Cell. 2013;13:382–384. [DOI] [PubMed] [Google Scholar]
- 15. Fairchild PJ. The challenge of immunogenicity in the quest for induced pluripotency. Nat Rev Immunol. 2010;10:868–875. [DOI] [PubMed] [Google Scholar]
- 16. Dierselhuis M, Goulmy E. The relevance of minor histocompatibility antigens in solid organ transplantation. Curr Opin Organ Transplant. 2009;14:419–425. [DOI] [PubMed] [Google Scholar]
- 17. Ingulli E. Mechanism of cellular rejection in transplantation. Pediatr Nephrol. 2010;25:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Daadi MM, Barberi T, Shi Q, Lanford RE. Nonhuman primate models in translational regenerative medicine. Stem Cells Dev. 2014;23(suppl 1):83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Messaoudi I, Estep R, Robinson B, Wong SW. Nonhuman primate models of human immunology. Antioxid Redox Signal. 2011;14:261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kean LS, Singh K, Blazar BR, Larsen CP. Nonhuman primate transplant models finally evolve: detailed immunogenetic analysis creates new models and strengthens the old. Am J Transplant. 2012;12:812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Phillips KA, Bales KL, Capitanio JP, Conley A, Czoty PW, ‘t Hart BA, Hopkins WD, Hu SL, Miller LA, Nader MA, Nathanielsz PW, Rogers J, Shively CA, Voytko ML. Why primate models matter. Am J Primatol. 2014;76:801–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wiseman RW, Karl JA, Bohn PS, Nimityongskul FA, Starrett GJ, O'Connor DH. Haplessly hoping: macaque major histocompatibility complex made easy. ILAR J. 2013;54:196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wiseman RW, Wojcechowskyj JA, Greene JM, Blasky AJ, Gopon T, Soma T, Friedrich TC, O'Connor SL, O'Connor DH. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex‐identical cynomolgus macaques from Mauritius. J Virol. 2007;81:349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bach F, Hirschhorn K. Lymphocyte interaction: a potential histocompatibility test in vitro. Science. 1964;143:813–814. [DOI] [PubMed] [Google Scholar]
- 25. Treiman GS, Lawrence PF, Rockwell WB. Autogenous arterial bypass grafts: durable patency and limb salvage in patients with inframalleolar occlusive disease and end‐stage renal disease. J Vasc Surg. 2000;32:13–22. [DOI] [PubMed] [Google Scholar]
- 26. Mees BM, Robinson DR, Fell G, Chan AT. Radial artery bypass graft is a feasible and durable conduit for challenging infrainguinal revascularization: 17 years of Melbourne experience. Eur J Vasc Endovasc Surg. 2014;48:80–87. [DOI] [PubMed] [Google Scholar]
- 27. Zhao T, Zhang ZN, Westenskow PD, Todorova D, Hu Z, Lin T, Rong Z, Kim J, He J, Wang M, Clegg DO, Yang YG, Zhang K, Friedlander M, Xu Y. Humanized mice reveal differential immunogenicity of cells derived from autologous induced pluripotent stem cells. Cell Stem Cell. 2015;17:353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Erlich HA, Opelz G, Hansen J. HLA DNA typing and transplantation. Immunity. 2001;14:347–356. [DOI] [PubMed] [Google Scholar]
- 29. Garces JC, Giusti S, Staffeld‐Coit C, Bohorquez H, Cohen AJ, Loss GE. Antibody‐mediated rejection: a review. Ochsner J. 2017;17:46–55. [PMC free article] [PubMed] [Google Scholar]
- 30. Shimizu K, Sugiyama S, Aikawa M, Fukumoto Y, Rabkin E, Libby P, Mitchell RN. Host bone‐marrow cells are a source of donor intimal smooth‐ muscle‐like cells in murine aortic transplant arteriopathy. Nat Med. 2001;7:738–741. [DOI] [PubMed] [Google Scholar]
- 31. Schwartz SM. Perspectives series: cell adhesion in vascular biology: smooth muscle migration in atherosclerosis and restenosis. J Clin Invest. 1997;99:2814–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DeRuiter MC, Poelmann RE, VanMunsteren JC, Mironov V, Markwald RR, Gittenberger‐de Groot AC. Embryonic endothelial cells transdifferentiate into mesenchymal cells expressing smooth muscle actins in vivo and in vitro. Circ Res. 1997;80:444–451. [DOI] [PubMed] [Google Scholar]
- 33. Frid MG, Kale VA, Stenmark KR. Mature vascular endothelium can give rise to smooth muscle cells via endothelial‐mesenchymal transdifferentiation: in vitro analysis. Circ Res. 2002;90:1189–1196. [DOI] [PubMed] [Google Scholar]
- 34. Sata M, Saiura A, Kunisato A, Tojo A, Okada S, Tokuhisa T, Hirai H, Makuuchi M, Hirata Y, Nagai R. Hematopoietic stem cells differentiate into vascular cells that participate in the pathogenesis of atherosclerosis. Nat Med. 2002;8:403–409. [DOI] [PubMed] [Google Scholar]
- 35. Religa P, Bojakowski K, Maksymowicz M, Bojakowska M, Sirsjo A, Gaciong Z, Olszewski W, Hedin U, Thyberg J. Smooth‐muscle progenitor cells of bone marrow origin contribute to the development of neointimal thickenings in rat aortic allografts and injured rat carotid arteries. Transplantation. 2002;74:1310–1315. [DOI] [PubMed] [Google Scholar]
- 36. Hillebrands JL, Klatter FA, Rozing J. Origin of vascular smooth muscle cells and the role of circulating stem cells in transplant arteriosclerosis. Arterioscler Thromb Vasc Biol. 2003;23:380–387. [DOI] [PubMed] [Google Scholar]
- 37. Mitchell RN, Libby P. Vascular remodeling in transplant vasculopathy. Circ Res. 2007;100:967–978. [DOI] [PubMed] [Google Scholar]
- 38. Hillebrands JL, Klatter FA, van den Hurk BM, Popa ER, Nieuwenhuis P, Rozing J. Origin of neointimal endothelium and alpha‐actin‐positive smooth muscle cells in transplant arteriosclerosis. J Clin Invest. 2001;107:1411–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grimm PC, Nickerson P, Jeffery J, Savani RC, Gough J, McKenna RM, Stern E, Rush DN. Neointimal and tubulointerstitial infiltration by recipient mesenchymal cells in chronic renal‐allograft rejection. N Engl J Med. 2001;345:93–97. [DOI] [PubMed] [Google Scholar]
- 40. Glaser R, Lu MM, Narula N, Epstein JA. Smooth muscle cells, but not myocytes, of host origin in transplanted human hearts. Circulation. 2002;106:17–19. [DOI] [PubMed] [Google Scholar]
- 41. Quaini F, Urbanek K, Beltrami AP, Finato N, Beltrami CA, Nadal‐Ginard B, Kajstura J, Leri A, Anversa P. Chimerism of the transplanted heart. N Engl J Med. 2002;346:5–15. [DOI] [PubMed] [Google Scholar]
- 42. McDonald AI, Shirali AS, Aragon R, Ma F, Hernandez G, Vaughn DA, Mack JJ, Lim TY, Sunshine H, Zhao P, Kalinichenko V, Hai T, Pelegrini M, Ardehali R, Iruela‐Arispe ML. Endothelial regeneration of large vessels is a biphasic process driven by local cells with distinct proliferative capacities. Cell Stem Cell. 2018;23:210–225.e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pashneh‐Tala S, MacNeil S, Claeyssens F. The tissue‐engineered vascular graft: past, present, and future. Tissue Eng Part B Rev. 2015;22:68–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roh JD, Sawh‐Martinez R, Brennan MP, Jay SM, Devine L, Rao DA, Yi T, Mirensky TL, Nalbandian A, Udelsman B, Hibino N, Shinoka T, Saltzman WM, Snyder E, Kyriakides TR, Pober JS, Breuer CK. Tissue‐engineered vascular grafts transform into mature blood vessels via an inflammation‐mediated process of vascular remodeling. Proc Natl Acad Sci USA. 2010;107:4669–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matsumura G, Hibino N, Ikada Y, Kurosawa H, Shin'oka T. Successful application of tissue engineered vascular autografts: clinical experience. Biomaterials. 2003;24:2303–2308. [DOI] [PubMed] [Google Scholar]
- 46. Le Blanc K, Tammik C, Rosendahl K, Zetterberg E, Ringdén O. HLA expression and immunologic properties of differentiated and undifferentiated mesenchymal stem cells. Exp Hematol. 2003;31:890–896. [DOI] [PubMed] [Google Scholar]
- 47. Reynolds AJ, Lawrence C, Cserhalmi‐Friedman PB, Christiano AM, Jahoda CAB. Trans‐gender induction of hair follicles. Nature. 1999;402:33. [DOI] [PubMed] [Google Scholar]
- 48. Wystrychowski W, McAllister TN, Zagalski K, Dusserre N, Cierpka L, L'Heureux N. First human use of an allogeneic tissue‐engineered vascular graft for hemodialysis access. J Vasc Surg. 2014;60:1353–1357. [DOI] [PubMed] [Google Scholar]
- 49. Vermeersch P, Agostoni P, Verheye S, Van den Heuvel P, Convens C, Bruining N, Van den Branden F, Van Langenhove G. Randomized double‐blind comparison of sirolimus‐eluting stent versus bare‐metal stent implantation in diseased saphenous vein grafts: six‐month angiographic, intravascular ultrasound, and clinical follow‐up of the RRISC Trial. J Am Coll Cardiol. 2006;48:2423–2431. [DOI] [PubMed] [Google Scholar]
- 50. Dahl SLM, Kypson AP, Lawson JH, Blum JL, Strader JT, Li Y, Manson RJ, Tente WE, DiBernardo L, Hensley MT, Carter R, Williams TP, Prichard HL, Dey MS, Begelman KG, Niklason LE. Readily available tissue‐engineered vascular grafts. Sci Transl Med. 2011;3:68ra69. [DOI] [PubMed] [Google Scholar]
- 51. Ogawa LM, Vallender EJ. Genetic substructure in cynomolgus macaques (Macaca fascicularis) on the island of Mauritius. BMC Genom. 2014;15:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hochberg EP, Miklos DB, Neuberg D, Eichner DA, McLaughlin SF, Mattes‐Ritz A, Alyea EP, Antin JH, Soiffer RJ, Ritz J. A novel rapid single nucleotide polymorphism (SNP)‐based method for assessment of hematopoietic chimerism after allogeneic stem cell transplantation. Blood. 2003;101:363–369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Autograft Patency
Table S2. Donor Specific Antibody (DSA)
Figure S1. Pre‐transplant donor‐specific antibodies. Representative graph summarizing the pre‐transplant results of one experiment showing DSA positive pair Cy0700 and Cy0673.
Figure S2. Assessment of cell identity within allografts. At end of study, radial artery grafts were explanted. The proximal and distal anastomosis sites were saved for sectioning and staining while the middle of the graft was processed and analyzed for the presence of host/donor cells. A, The graft was digested and individual graft cells were sorted for CD31+ and CD31‐ populations. Shown is a representative sort for CD31 cells. B, Plot of SNP analysis results from all samples in the study. C, Plot of SNP analysis results from a partial haplomatch (left panel) and complete mismatch animal (right panel) that showed a mixture host and donor endothelial cells within the graft.


