FIGURE 2:
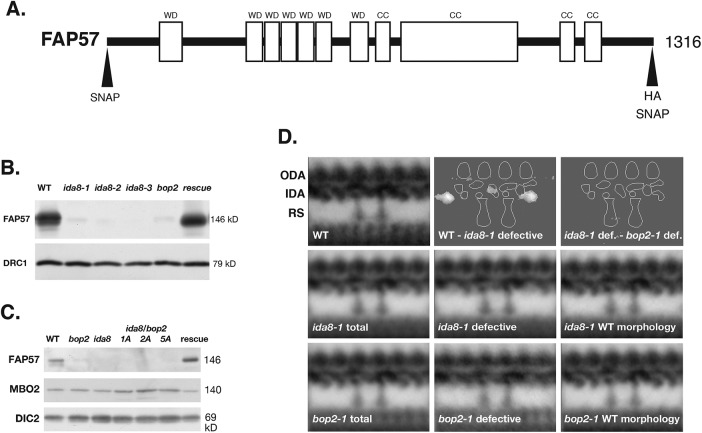
The ida8 and bop2 mutants have similar biochemical and structural defects. (A) Diagram of the IDA8 gene product, FAP57, showing the location of the predicted WD repeat domains (WD) in the first half of the polypeptide and the coiled coil domains (CC) in the second half. Also shown are the positions of the HA and SNAP tags at the N-terminal and C-terminal ends. (B) Western blot of axonemes from WT, three ida8 mutants, bop2-1, and an ida8-1; FAP57 strain (CC-4486, rescued with the 6h9 Bac clone) was probed with antibodies against FAP57 and the N-DRC subunit DRC1 as a loading control. (C) Western blot of axonemes from WT, bop2-1, ida8-1, three ida8-1/bop2-1 diploids (1A, 2A, 5A), and an ida8-1; FAP57 strain (CC-4486) was probed with antibodies against FAP57, MBO2, and the outer arm dynein IC2 (DIC2) subunit, also known as IC69, as a loading control. (D) Averages and difference plots of the 96 nm repeat obtained by TEM of thin sections and image averaging as described in O’Toole et al. (1995). The WT grand average (top left) was obtained from six axonemes with 65 repeats. The approximate locations of the ODA, IDA, and two RS are indicated. The ida8-1 grand average in the second row (ida8-1 total) was obtained from 41 axonemes with 379 repeats. The ida8-1 averages were divided into two classes, those with defective morphology (25 axonemes with 237 repeats) and those with WT morphology (16 axonemes with 142 repeats). The bop2-1 grand average in the third row (bop2-1 total) was obtained from 22 axonemes with 196 repeats. These were also separated into two classes, bop2-1 defective (8 axonemes with 66 repeats) and bop2-1 with WT morphology (14 axonemes with 130 repeats). The difference plots in the top row show identified two densities in the 96 nm repeat that were significantly different (P < 0.05) between the WT and ida8-1 defective grand averages. No significant difference was detected between the ida8-1 defective average and bop2-1 defective average.