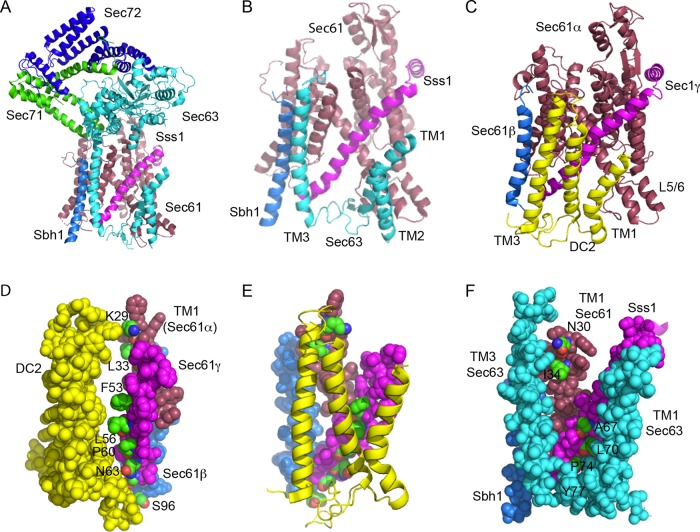
FIGURE 1:
Comparison of the Sec63p and DC2 binding sites on the yeast and mammalian Sec61 complexes. (A) Structure of the yeast Sec complex. Sec61p is raspberry, Sbh1p is marine, Sss1 is magenta, Sec63p is cyan, Sec71p is green, and Sec72p is blue. (B) Enlarged view of the interactions between the three TMs of Sec63p and the Sec61 heterotrimer, using the same color code as in panel A. The cytoplasmic domains of Sec63p, Sec71p, and Sec72p have been removed for clarity. The cytosolic (L1/2) and lumenal (L2/3) loops of Sec63p were not completely resolved. (C) Interaction between DC2 (yellow) and the mammalian Sec61 heterotrimer (color codes as in A). The other OST subunits are omitted for clarity, as the STT3A protein binds to the opposite face of DC2 from the Sec61 complex. (D) Space-filling model showing the interaction surface of DC2 and the TM spans of Sec61β, Sec61γ, and TM1 of Sec61α. Proteins are color-coded as in C. Residues in the Sec61 complex that compose the interaction surface (K29 and L33 in Sec61α, F53, L56, P60, and N63 in Sec61γ, and S96 in Sec61β) are color-coded by element and are labeled. (E) DC2-Sec61 complex interaction surface color-coded as in D. (F) Space-filling model of the interface between Sec63p (cyan; TM1, TM3, and the resolved section of L2), Sss1p (magenta), Sbh1p (slate), and TM1 of Sec1p (raspberry). The residues in the yeast Sec61 complex subunits that align with residues shown in panels D and E (Supplemental Table S1) are color-coded by element and are labeled. P74 and Y77 in Sss1p are completely buried by Sec63p, while other interaction residues are partially buried. The figure was made with PYMOL v2.1 software and PDB files 6N3Q (A, B, F) and 6FTI (C–E).