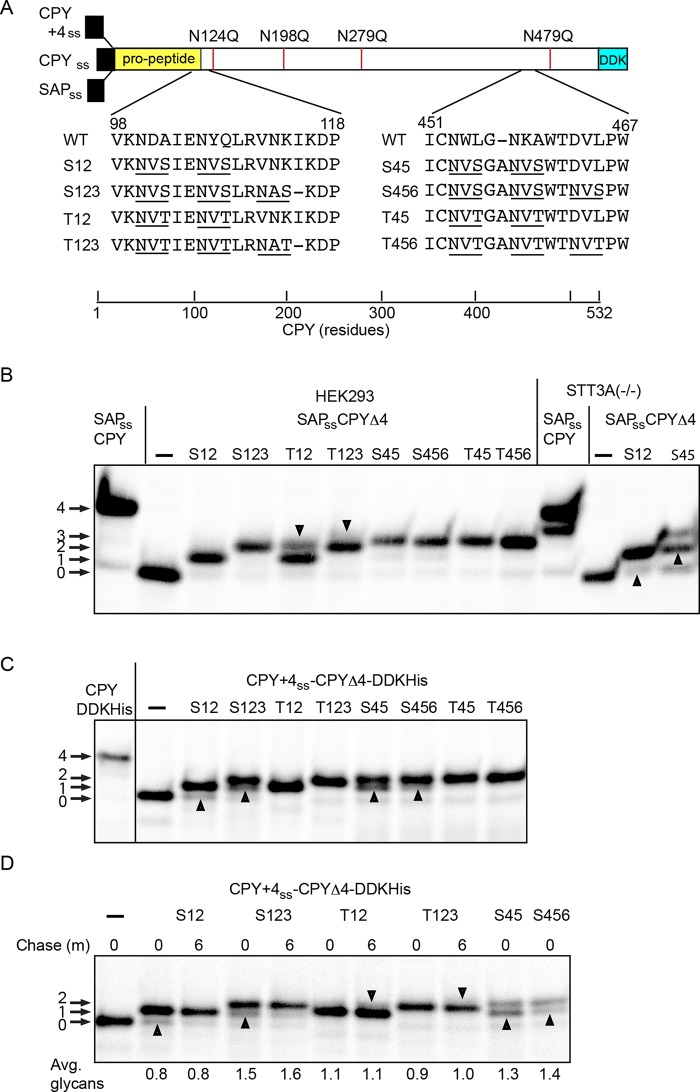
FIGURE 5:
Glycosylation of CPY derivatives in yeast and human cells. (A) Diagram of CPY derivatives with closely spaced acceptor sites. The wild-type signal sequence of CPY was replaced with either the prosaposin signal sequence (SAPss) for expression in human cells or the CPY+4 sequence to direct cotranslational translocation in yeast. The four glycosylation sites in CPY were eliminated by the indicated N to Q mutations to create the SAPss-CPYΔ4 and CPY+4ss-CPYΔ4 constructs. Two or three closely spaced NXS or NXT sites were created at the indicated sites to yield eight reporters. The introduced glycosylation sites are underlined. (B) Pulse labeling of the SAPss-CPY and SAPss-CPYΔ4 derivatives in wild-type and STT3A(–/–) HEK293 cells. Downward-pointing arrowheads indicate the fully glycosylated form of the T12 and T123 reporters. Upward-pointing arrowheads indicate hypoglycosylated forms of S12 and S45 reporter that were detected in STT3A(–/–) cells. (C) Pulse labeling (4 min) of CPY+4ss-CPY and CPY+4ss-CPYΔ4 derivatives expressed in yeast. Upward-pointing arrowheads designate hypoglycosylated forms of the reporters that are either less abundant or not detected in wild-type HEK293 cells. Note the absence of the fully glycosylated T12 and T123 reporters. (D) Pulse–chase labeling of selected CPY+4ss-CPYΔ4 derivatives in yeast. The pulse period was 2 min for all samples. Upward-pointing arrowheads designate hypoglycosylated forms of NXS containing reporters that are more prominent before the chase period. The downward-pointing arrowheads designate trace amounts of the fully glycosylated T12 and T123 reporters that were visible after the chase. The quantified values below the gel lanes are for the experiment shown, which is representative of two similar experiments.