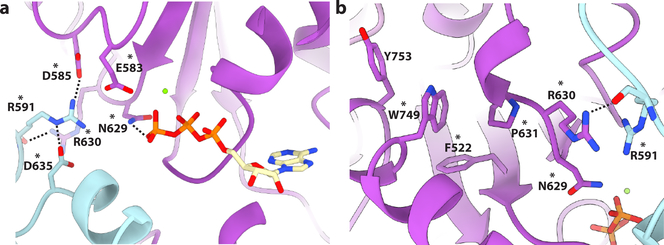
Figure 5. An allosteric network mutated in HSP couples substrate binding to oligomerization and ATP hydrolysis.
(a) Interaction network centered on R591. Residues colored plum and cyan for protomers B and C, respectively. Residues mutated in HSP are indicated with an asterix. All interactions depicted have a measured distance of less than 4 Å. (b) W749 makes van der Walls contacts with several hydrophobic residues mutated in HSP and connects pore loop 3 to the C-terminal helix α11 involved in oligomerization. Residues are colored purple and cyan for protomers C and D, respectively. Residues mutated in HSP are indicated with an asterix. All interactions depicted have a measured distance of less than 4 Å.