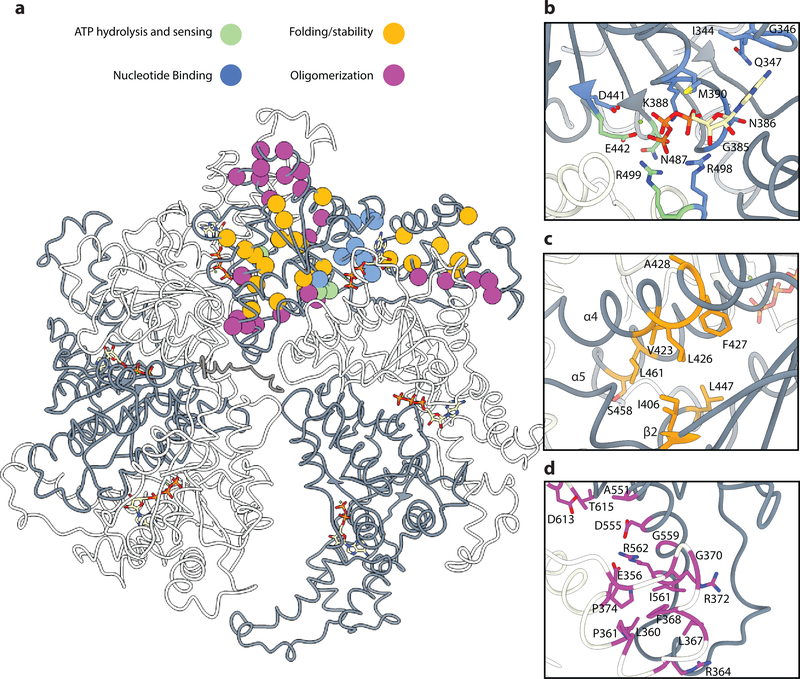
Figure 6. Structure-based insight into the mechanism of action of human spastin HSP disease mutations.
(a) Homology model of the human spastin hexamer depicting the location of missense mutations identified in HSP. Protomers are colored in alternating grey and white around the hexamer. Mutations are depicted as spheres at their Cα position and colored according to their proposed mechanism of action: interference with nucleotide binding (blue), defects in ATP hydrolysis and γ-phosphate sensing (green), disruption of interactions important for folding or conferring stability to the protomer (orange), and disruption of oligomerization interfaces (magenta). (b) Close-up view of the nucleotide pocket, showing a large number of mutations that likely disrupt nucleotide binding or impair ATP hydrolysis and sensing. (c) Close-up view of hydrophobic interfaces between α4, α5 and β2 highlighting potentially destabilizing HSP mutations. (d) Close-up view of the NBD-HBD interaction interface between protomers showing a high-density of HSP mutations.