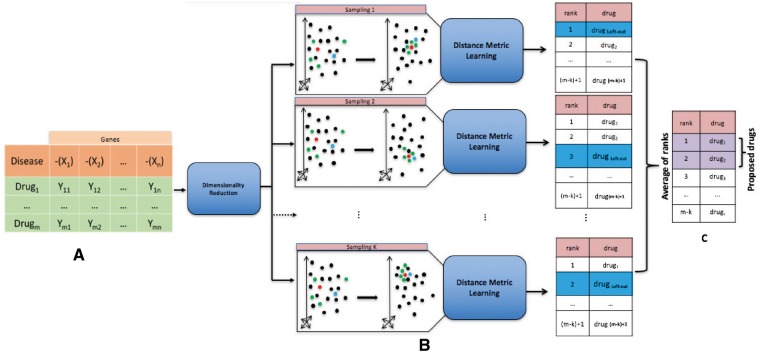
Fig. 1.
Framework overview. (A) We transform the input matrix into a lower dimensionality matrix by applying LLE (Roweis and Saul, 2000). (B) We incorporate the known relationship between disease and its FDA-approved drugs into this space using DML algorithm. Our hypothesis is that under this new space and the new metric, the clinically relevant drugs get close to the disease. We use the leave-one-out cross validation. In particular, in each round of sampling, we use one of the FDA-approved drugs for the disease (called left-out drug) for validation and we use the rest of them for training the DML algorithm. We then rank the list of drugs from the nearest one to the disease gene expression profile to the farthest one, based on the new learned metric. We repeat this procedure k times (where k is the number of FDA-approved drugs for the disease). (C) For each drug, an average of ranks is calculated over the k rounds of sampling. The proposed drugs are the ones with the lower average rank. In this figure, the red point represents the disease, the blue point represents the left-out drug and the green point represents the FDA-approved drug for the disease used for training the DML algorithm