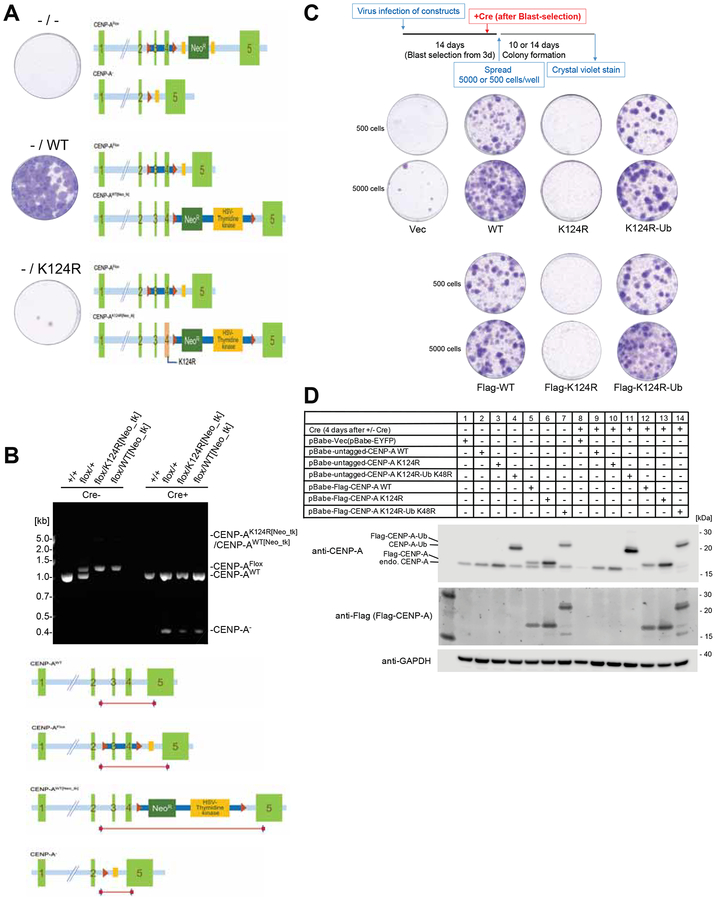
Figure 4. The hemizygous CENP-A K124R knock-in mutant created by the CRISPR-Cas9 system is lethal.
(A) The hemizygous K124R mutant is lethal to cells. Representative images from the colony outgrowth assay for cells harboring the modified CENP-A genomic locus shown as schematic illustration. Positions of the CENP-A exons, loxP sites, FRT sites, and the neomycin-resistant gene-thymidine kinase gene cassette are indicated. Images of colonies visualized with crystal violet were taken 14 days after infection with Cre expressing retrovirus vector.
(B) CENP-A genotypes validated by genomic PCR. Genomic structures of CENP-Aflox/K124R[Neo_tk] cells and its wildtype counterpart CENP-Aflox/WT[Neo_tk] cells were confirmed by genomic PCR using the specific primer sets. Genomic DNA was isolated from Wild type (+/+), Parental strain (flox/+), CENP-Aflox/K124R[Neo_tk] (flox/K124R[Neo_tk]) and CENP-Aflox/WT[Neo_tk] (flox/WT[Neo_tk]) before (Cre-) and 4 days after infection with Cre expressing retrovirus (Cre+). The schematic illustration indicates the positions of PCR products.
(C) Viability of Flag-tagged/untagged CENP-AK124R mutants is recovered by mono-ubiquitin fusion. Representative images from the colony outgrowth assay as shown in schematic (top) of different conditions (See STAR METHODS). Vec: pBabe-EYFP; WT: pBabe-CENP-A; K124R: pBabe-CENP-A K124R; K124R-Ub: pBabe-CENP-A K124R-Ub K48R; Flag-WT: pBabe-Flag-CENP-A; Flag-K124R: pBabe-Flag-CENP-A K124R; Flag-K124R-Ub: pBabe-Flag-CENP-A K124R-Ub K48R.
(D) Confirmation of expression levels of the indicated rescue constructs and endogenous CENP-A protein (ca. 16 kDa) before and 4 days after Cre infection. GAPDH protein was used as a loading control.