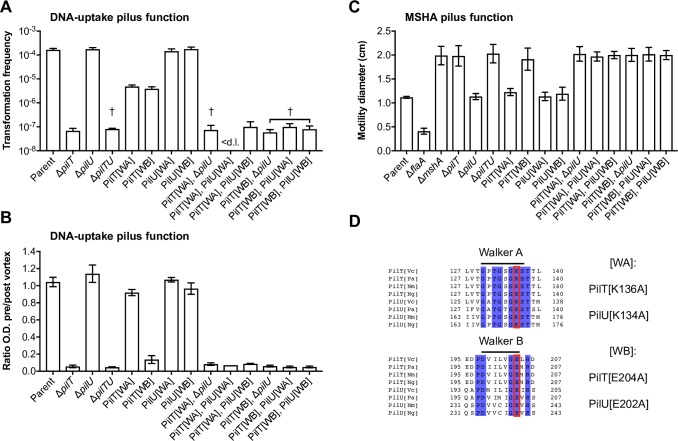
Fig 1. Inactivated PilT variants reveal a role for PilU in DNA-uptake pilus and MSHA pilus function.
(A-C) The role of PilT and PilU, and their corresponding Walker A and Walker B variants, was assessed using (A) natural transformation and (B) auto-aggregation as readouts for DNA-uptake pilus function, and (C) by swarming motility as a readout for MSHA pilus function. All strains contain an arabinose-inducible copy of tfoX (araC PBAD-tfoX; TntfoX), within an ectopically integrated transposon, which was used for chitin-independent competence induction in A and B. (A) Chitin-independent transformation assay. Transformation frequencies are the mean of three repeats (+S.D.). < d.l., below detection limit. †, < d.l. in one repeat. (B) Aggregation is shown as the ratio of the culture optical density (O.D. 600 nm) before and after vortexing in the presence of tfoX induction. Values are the mean of three repeats (±S.D.). Values close to 1 = No aggregation. Values close to 0 = Full aggregation. (C) Surface motility was determined on soft LB agar plates. The swarming diameter (cm) is the mean of three repeats (±S.D.). A flagellin-deficient (ΔflaA) non-motile strain was used as a negative control. (D) Alignments highlighting the conserved Walker A and Walker B motifs of PilT and PilU. Protein sequences were aligned using Clustal Omega and the figure prepared using Jalview. Identical residues are shaded in blue. Residues targeted for mutagenesis in the Walker A motif (i.e. PilT[K136] and PilU[K134]) and the atypical Walker B motif (i.e. PilT[E204] and PilU[E202]) are boxed in red. Species abbreviations: [Vc]; Vibrio cholerae, [Pa]; Pseudomonas aeruginosa, [Nm]; Neisseria meningitidis, [Ng]; Neisseria gonorrhoeae.