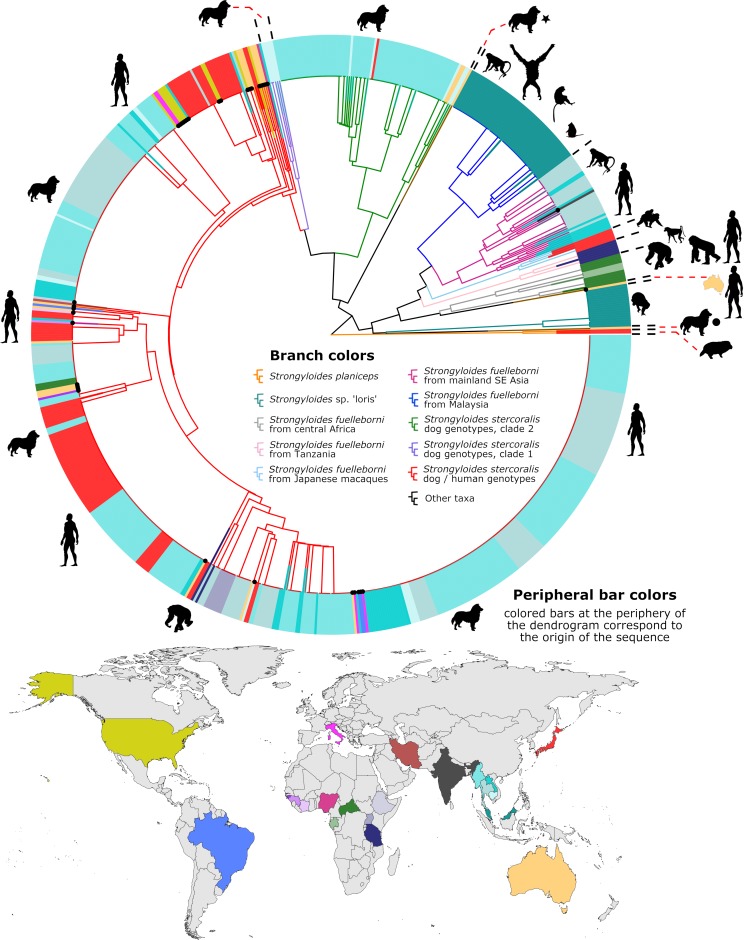
Fig 2. Dendrogram of clustered cox1 sequences.
This dendrogram represents 787 cox1 sequences, including those generated in this study (branches tipped in a black dot) and all published cox1 sequences from GB that overlap completely with our 217 base cox1 amplicon (to our knowledge). Peripheral bars are colored according to their site of origin, which corresponds to the colored countries on the map. Branches are color coded separately, according to their identity; either a species assignment, a genus, or their S. stercoralis genotype. The dog image with a black star indicates a sequence from an Australian dog generated by us previously [21], that is distinct from other Strongyloides spp. and clusters between the S. stercoralis and S. fuelleborni groups. The dog image with a black circle highlights a published sequence [21] that clusters close to, yet is distinct from Strongyloides spp. detected previously in lorises [27]. Animal images reflect the mammalian hosts that the sequences were associated with. The miniaturized image of Australia next to a human silhouette shows the location of a unique S. fuelleborni cox1 sequence. Two sequences of Strongyloides planiceps (orange branches) from Japanese raccoon dogs serve as an outgroup. The identity of each sequence is provided in S1 Fig, which is a searchable PDF of the same dendrogram with all GB accession numbers, the countries of origin, and host species provided. The GB accession numbers for sequences in this dendrogram that were generated as part of this study (branches tipped in a black dot) are provided in S1 File. The sequences used to construct this dendrogram are provided in S2 File.