Fig. 8.
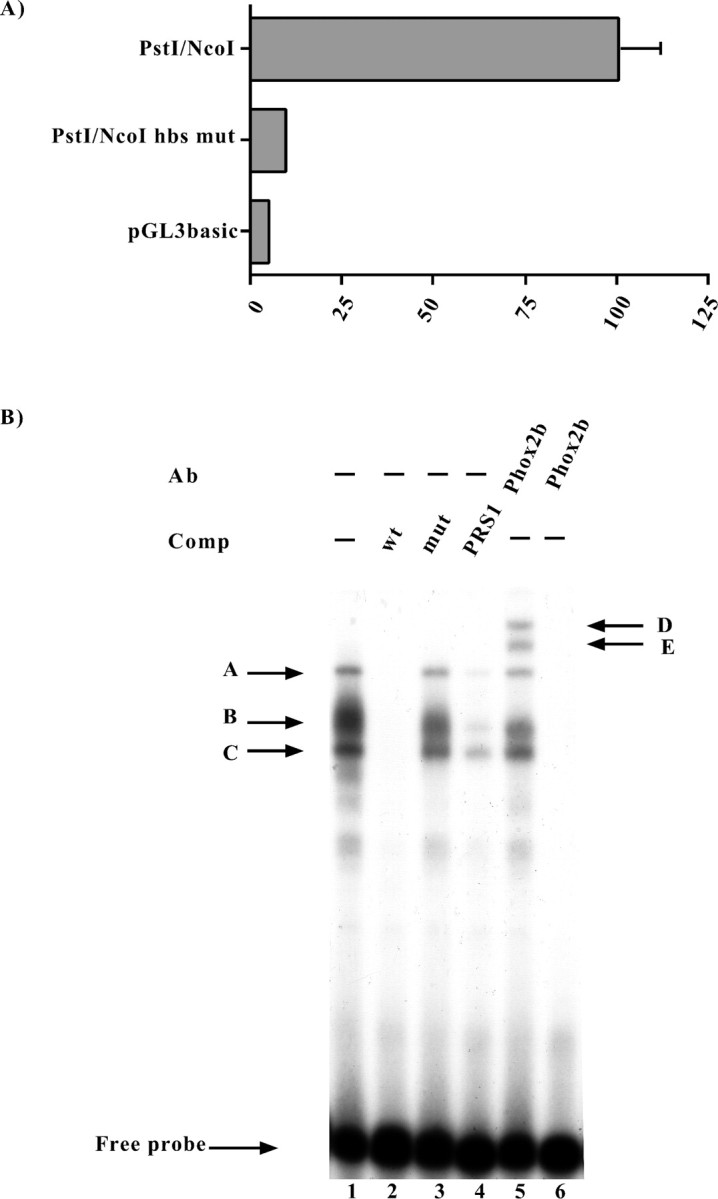
Functional and molecular characterization of the putative homeoprotein binding site (hbs).A, Luciferase assays. SY5Y cells were transiently transfected with the constructs indicated on the left, and the luciferase assays were performed 48 hr later. Thebars represent the transcriptional activity of the constructs given as the percentage of the activity of thePstI/NcoI construct. The data represent the means ± SE (error bars) of at least three independent experiments performed in duplicate. B, EMSAs were performed using a probe corresponding to the human Phox2a sequence between the nucleotides −363 and −337 (Fig. 2), which contained the putative homeoprotein binding site. On the left, thearrows and corresponding letters(A, B, C) indicate the retarded complexes that formed on incubation of the labeled probe with the SY5Y nuclear extract (lane 1). Competition experiments were performed in the presence of a 1000× excess of the wild-type oligonucleotide (lane 2) or in the presence of its mutated version in which the putative homeoprotein binding site was mutagenized (lane 3). The specificity of the DNA–protein interactions was also checked by means of competition with a cold oligonucleotide bearing the Phox2 site of thedopamine-β-hydroxylase gene (Yang et al., 1998) (lane 4). On the right, the arrows and corresponding letters(D, E) indicate the supershifted complexes obtained using an antibody directed against Phox2b (Pattyn et al., 1997) (lane 5). Lane 6, No nuclear extract.
