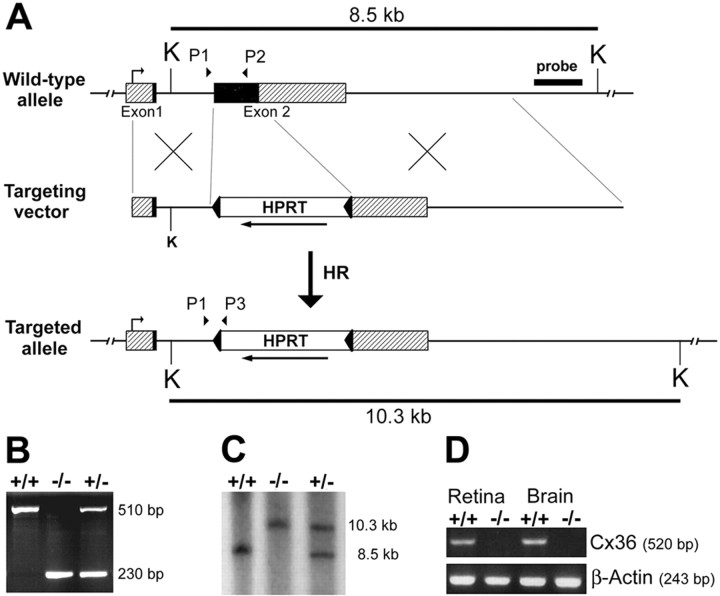
Fig. 1.
Generation of connexin36-deficient mice.A, Targeting scheme. The coding region of the wild-type Cx36 gene consists of two exons (boxes) that are separated by a 1100 bp intron. Exon1 contains a 530 bp noncoding region (hatched) and a 70 bp coding region (black). Exon 2 consists of a 900 bp coding region (black) and a 1400 bp noncoding region (hatched). After electrotransfecting the linearized targeting vector into HM-1 ES cells, homologous recombination occurred in 3% of all clones analyzed. In the mutated Cx36 allele, the complete coding region of exon2 and 170 bp of the flanking noncoding region were replaced by an HPRT minigene that was flanked by loxP sites (arrowheads) and transcribed in opposite direction (arrow) to Cx36. B, Diagnostic PCR of tail-cut DNA was performed to screen pedigree mice for the targeting effort. PCR was established using a three primer approach with a Cx36 intron-specific upstream primer (P1) and either a Cx36 exon2-specific downstream primer (P2), resulting in a 510 bp wild-type amplicon, or an HPRT-specific downstream primer (P3), giving a 230 bp amplicon for the mutated allele.C, Southern blot analysis was used to confirm PCR screening. KpnI-digested (K) tail-cut DNA was probed with an 860 bp externalXhoI/HindIII fragment. The smaller fragment (8.5 kb) resulted from the wild-type allele, the larger band (10.3 kb) derived from the mutated allele. D, RT-PCR. cDNA preparations of wild-type and Cx36 −/− tissues were used to check for the presence of Cx36 transcripts. PCR using an intron-spanning primer pair yielded a 520 bp amplicon specific for Cx36 cDNA in wild-type retina and brain but not in the corresponding Cx36 −/− cDNA preparations. The use of equal amounts of cDNA was verified by a separate β-actin PCR that yielded a 243 bp amplicon.