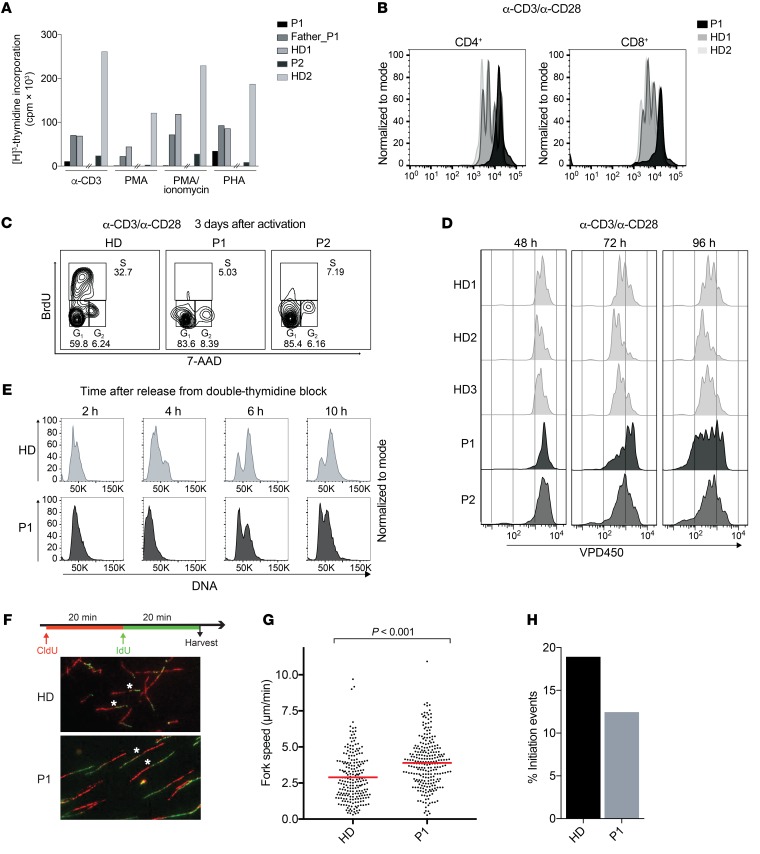
Figure 3. Polymerase δ –deficient cells show reduced proliferation, cell-cycle delay, and disturbed replication fork dynamics.
(A) T cell proliferation as measured by 48-hour thymidine incorporation upon stimulation of PBMCs with anti-CD3, PMA, PMA-ionomycin, and PHA. The hash marks separate independent experiments performed in P1 and P2. (B) T cell proliferation as measured by VPD450 three days after stimulation of PBMCs with anti-CD3 and anti-CD28. Results are representative of 2 independent experiments. (C) Cell-cycle analysis of patient and healthy control T cells as measured by BrdU incorporation. Data are representative of 2 independent experiments. (D) T cell proliferation as measured by VPD450 at the indicated time points after anti-CD3 and anti-CD28 stimulation of in vitro–expanded T cells. (E) Cell-cycle analysis of patient and healthy control fibroblasts upon double-thymidine block and release. (F) Scheme of dual-pulse labeling in DNA fiber analysis and representative images of the fiber analysis experiment performed using patient and HD fibroblasts. Original magnification, ×40. Representative green tracks are marked with an asterisk. Fiber data are representative of 2 independent experiments. Number of fibers counted: 1859 in HDs and 1213 in P1. (G) Replication fork speed in P1 (average of 3.899 μm/min) and HD (average of 2.89 μm/min) fibroblasts as measured by the length of IdU tracks. Statistical significance calculated by Wilcoxon rank sum test. (H) Analysis of initiation events in P1 (12.45%) and HD (18.93%) fibroblasts as measured by the frequency of only IdU-labeled tracks.