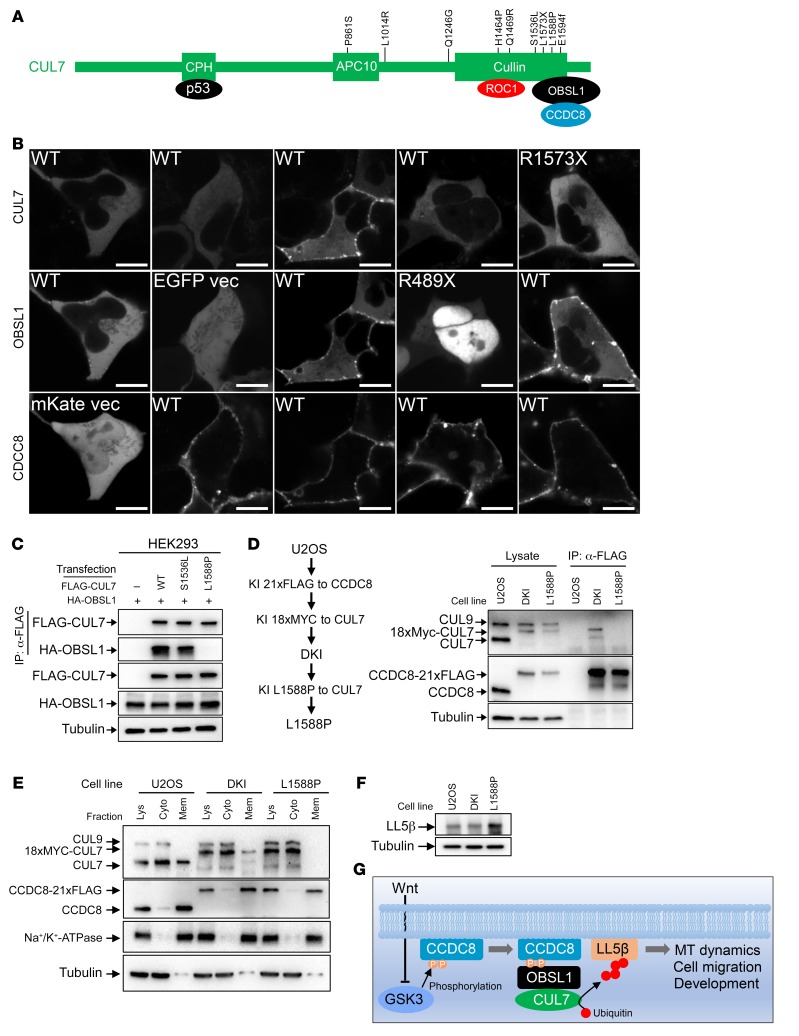
Figure 7. 3-M syndrome patient–derived mutations disrupt the assembly of the 3-M complex on the plasma membrane and the regulation of LL5β.
(A) Schematic representation of domains, binding proteins, and 3-M patient–derived mutations in CUL7. (B) mKate-fused wild-type or mutant CCDC8 was coexpressed with CUL7-mTagBFP and OBSL1-EGFP in U2OS cells, followed by confocal microscopic analyses of protein localization. Scale bars: 10 μm. (C) FLAG-CUL7 and HA-OBSL1 were coexpressed in HEK293 cells. CUL7-OBSL1 association was determined by IP and Western analysis. (D) U2OS cells with CCDC8-21×FLAG and 16×MYC-CUL7 double knockin (DKI, see Figure 2 for details) were subjected to CRISPR-mediated genome editing to introduce a homozygous L1588P mutation. The binding between endogenous CUL7 and CCDC8 was determined by IP and Western. (E) Total cell lysates derived from parental, DKI, and DKI/L1588P U2OS cells were fractionated by centrifugation, and total lysate (Lys), cytoplasm (Cyto), and membrane (Mem) fractions were analyzed by Western blotting. (F) The steady-state level of LL5β protein was determined by direct Western blot in parental, DKI, and DKI/L1588P U2OS cells. (G) Schematic representation of the function and regulation of 3-M E3 ubiquitin ligase. MT, microtubule.