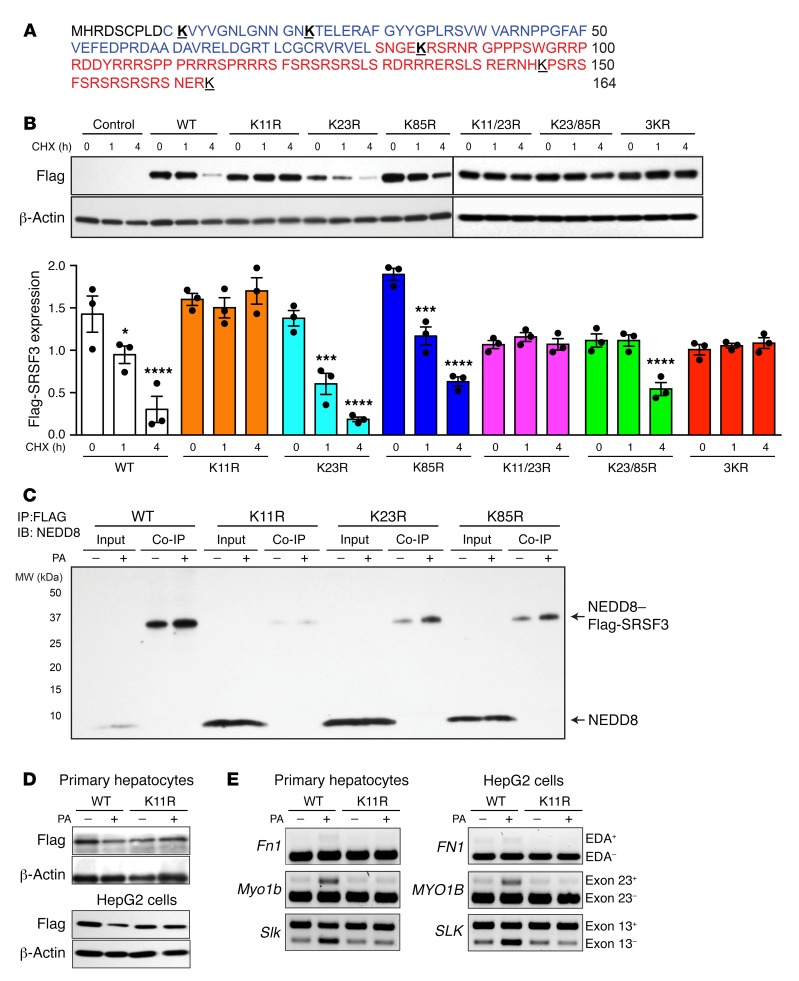
Figure 5. Mutation of lysine 11 prevented SRSF3 protein degradation.
(A) Amino acid sequence of SRSF3 protein. The RNA recognition motif (RRM) is shown in blue, and the arginine- and serine-rich (RS) domain in red. Lysine residues are bold face and underlined. (B) HepG2 cells were transfected with empty vector (Control), wild-type Flag-SRSF3 (WT), or a series of single lysine-to-arginine mutants (K11R, K23R, K85R), double lysine mutants (K11/23R, K23/85R), or a triple lysine mutant (3KR) as indicated. Cells were treated with cycloheximide (CHX) for 1 and 4 hours, and then lysates were immunoblotted using the anti-Flag antibody. Graphs below show quantification of Flag-SRSF3 levels from transfected HepG2 cells. Results are presented as mean ± SEM (n = 3/group). *P < 0.05, ***P < 0.001, ****P < 0.0001 vs. time 0 by 1-way ANOVA. Colors indicate the different mutants. (C) HepG2 cells were transfected with Flag-SRSF3 or single lysine mutants (K11R, K23R, K85R) as indicated and treated with 500 μM PA for 12 hours. Cell lysates were immunoprecipitated with anti-Flag antibodies and then immunoblotted for NEDD8. Input indicates input control, Co-IP indicates the coimmunoprecipitated complex. Monomeric NEDD8 and the NEDD8–Flag-SRSF3 conjugate are indicated by arrows. (D) Analysis of PA-induced SRSF3 degradation. Primary hepatocytes (top panels) and HepG2 cells (bottom panels) were transfected with Flag-SRSF3-WT or Flag-SRSF3-K11R. Twenty-four hours later, primary hepatocytes or HepG2 cells were treated with 250 μM or 500 μM PA, respectively, for 12 hours. Cells were then lysed and immunoblotted with the anti-Flag antibody. (E) Analysis of PA-induced changes in RNA splicing. Primary hepatocytes (left panels) and HepG2 cells (right panels) were transfected with Flag-SRSF3-WT or Flag-SRSF3-K11R. Twenty-four hours later, cells were treated with 250 μM or 500 μM PA, respectively, for 12 hours. Exon 33 (EDA exon) incorporation in the Fn1 mRNA, Slk–exon 13 skipping, and Myo1b–exon 23 inclusion was analyzed by RT-PCR.