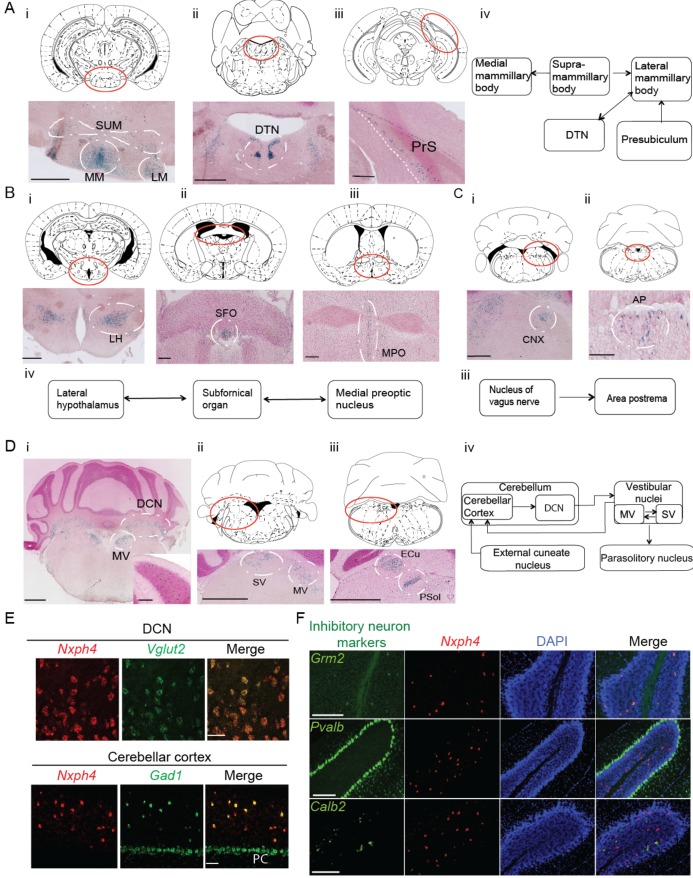
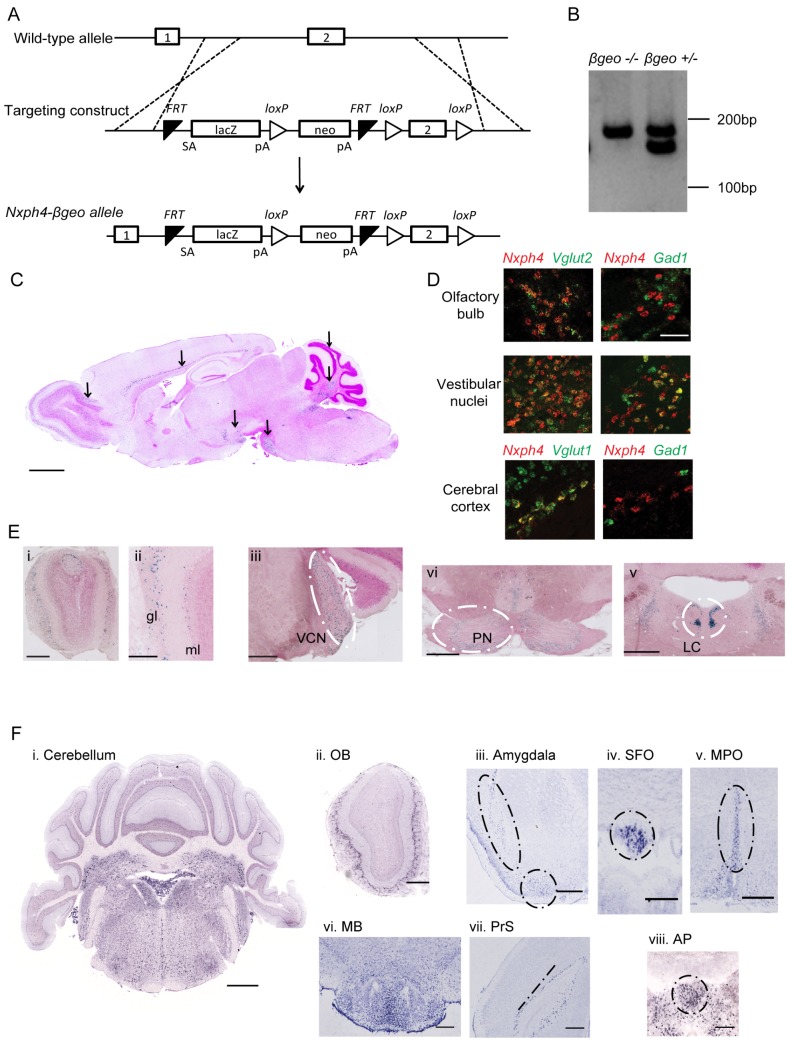
Figure 1. Nxph4 expression marks the components of select brain circuits.
(A–D) β-galactosidase staining of adult Nxph4βgeo/+ mice shows signals in mammillary body-related circuits (A), circumventricular organs (B–C), and cerebellar-vestibular circuits (D). Blue staining represents β-galactosidase activity. Top panels are stereotaxic maps adapted from the Paxinos and Franklin mouse brain atlas, with the red circle indicating the region for the image shown on the bottom panels. Aiv, Biv, Ciii, and Div illustrate the main connections among Nxph4+ regions in each circuit described. The inset in Di shows the blue staining in the cerebellar cortex granular layer. Scale bars: Ai, 500 μm; Aii, 100 μm; Aiii, 500 μm; Bi 500 μm; Bii, Biii 200 μm; Ci, Cii 100 μm; Di, Dii, Diii, 1 mm; Di inset, 100 μm. SUM, supramammillary body; MM, medial mammillary body; LM, lateral mammillary body; DTN, dorsal tegmental nucleus; PrS, presubiculum; LH, lateral hypothalamus; SFO, subfornical organ; MPO, median preoptic nucleus; CNX, nucleus of vagus nerve; AP, area postrema; DCN, deep cerebellar nuclei; MV, medial vestibular nucleus; SV, superior vestibular nucleus; ECu, external cuneate nucleus; Psol, parasolitary nucleus. (E) Double in situ staining of adult wild type mouse DCN and cerebellar cortex with probes against Nxph4, Vglut2 (an excitatory neuron marker), and Gad1 (an inhibitory neuron marker). PC: Purkinje cells. Scale bars: 50 μm. (F) Double in situ staining of mouse cerebellum shows that Nxph4 signals overlap with Grm2 (a Golgi cell marker) but not Pvalb or Calb2. Scale bars: 200 μm.