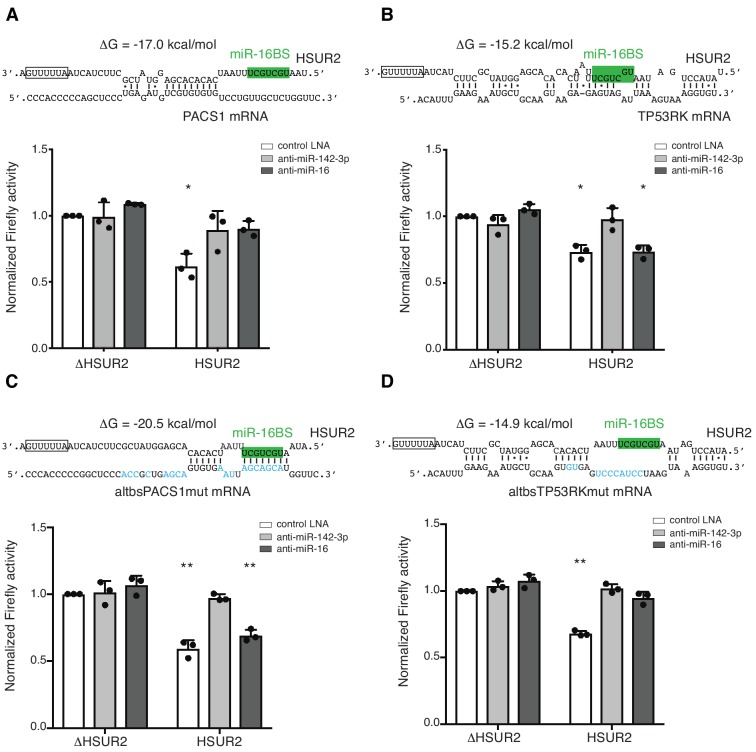
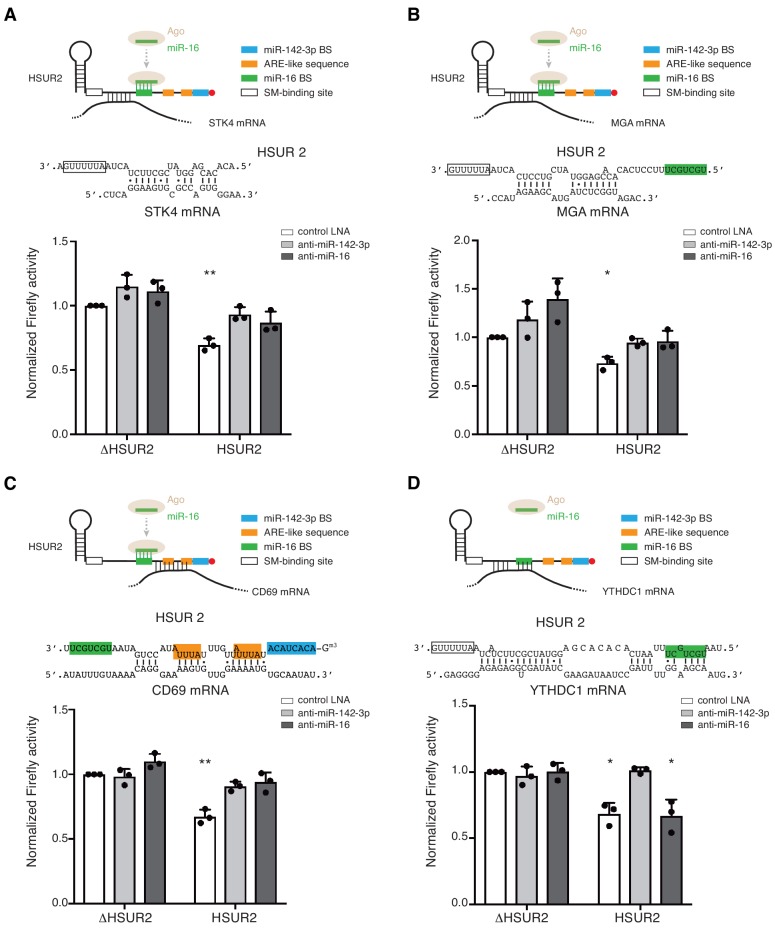
Figure 6. Base-pairing with target mRNA dictates the use of miR-16 in HSUR2-mediated mRNA repression.
(A) U937 cells stably expressing the Firefly luciferase gene with partial PACS1 3′UTR (Figure 2a) were transiently co-transfected with a plasmid carrying GFP and either HSUR2 promoter alone (ΔHSUR2) or wild-type HSUR2 (HSUR2) and a control LNA inhibitor, or an LNA inhibitor with complementarity to miR-142–3p or miR-16. (B–D) Same as in (A) but with cells stably expressing the Firefly luciferase gene with partial TP53RK 3′UTR (B), mutant PACS1 (altbsPACS1mut) 3′UTR (C), or mutant TP53RK (altbsTP53RKmut) 3′UTR (D). Modified residues are shown in blue. Dots represent mean values of independent experiments (n = 3) with error bars representing s.d. Two-sided one-sample Student’s t-test vs. control (ΔHSUR2) set at 1.0. *p<0.05, **p<0.01.