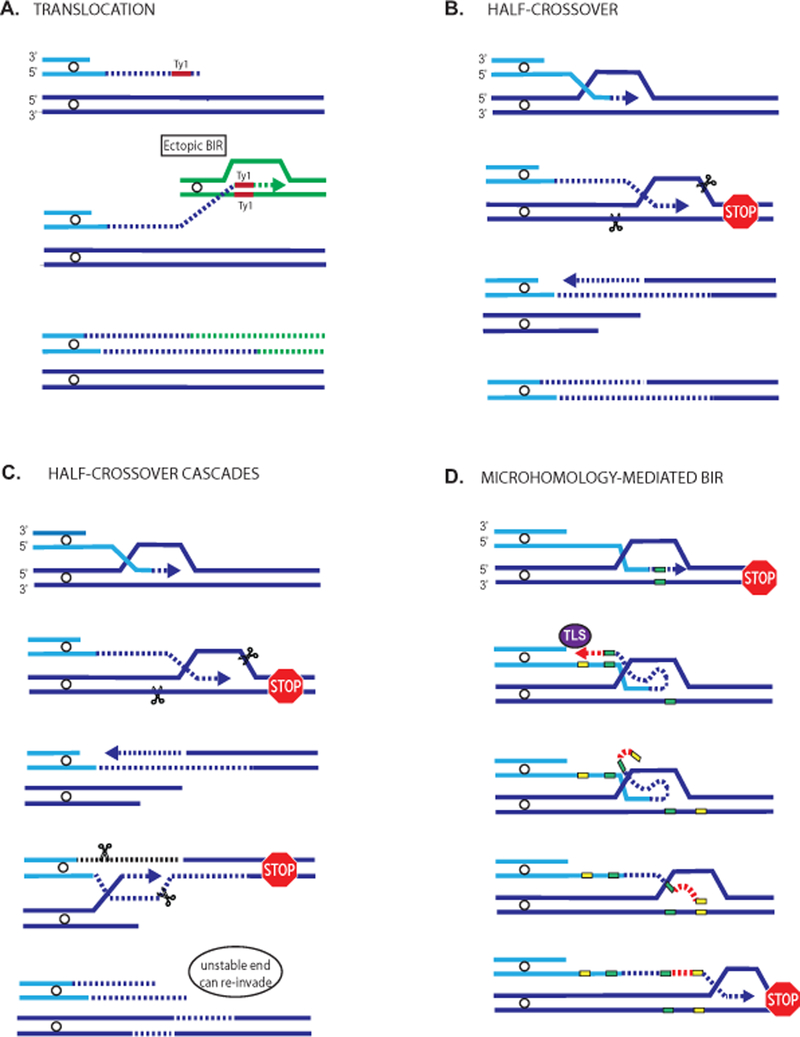
Figure 3:
Mechanisms of chromosomal rearrangements associated with BIR. A. A simple non-reciprocal translocation (NRT) resulting from long ssDNA resection at a DSB exposing repetitive DNA (Ty elements shown in red rectangles) that can initiate BIR at an ectopic position (non-homologous chromosome represented in green) through annealing of Ty elements. B. Formation of half-crossovers resulting from interruption of BIR that promotes a resolution of the migrating bubble, and resulting in fusion of the recombining portions of recipient and donor chromosome (shown in light blue and dark blue respectively). The remaining portion of the broken donor chromosome can either be lost as depicted in (B), or it can invade another donor at ectopic position (shown in green) leading to translocation (A). The invasion at ectopic position usually occurs at position of Ty or other repeated element. (C). The formation of half-crossover is followed by secondary BIR event resulting from the invasion of the broken donor into the half-crossover product, which can perpetuate breaking and re-invading cycles leading to half-crossover cascades (as described in (Vasan et al., 2014), or cycles of NRTs (not illustrated). D. Microhomology-mediated BIR resulting from a collapse of BIR synthesis resulting in dissociation of 3’ end from its template followed by annealing at microhomology in the region of ssDNA accumulated behind the BIR bubble. Using this annealing intermediate as a primer, TLS polymerases initiate DNA synthesis that eventually disrupts causing a second template switch by re-annealing to the original track of BIR through microhomology as described in (Sakofsky et al., 2015). Yellow and green boxes show regions of microhomology that can consist of 1–10bp. A color version of the figure is available online.