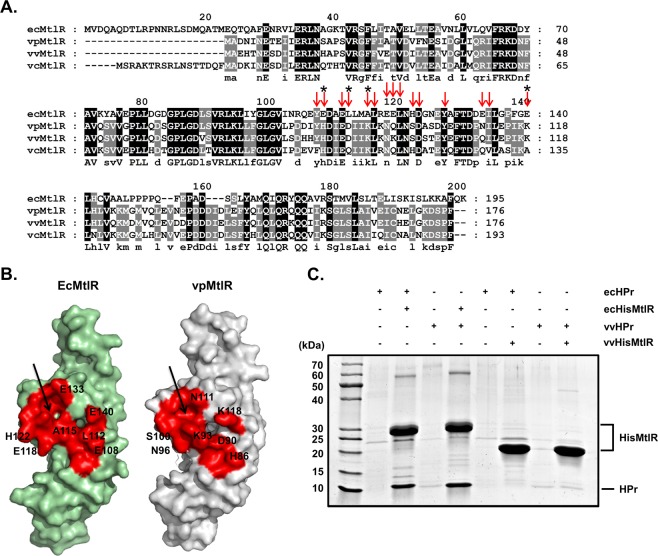
Figure 5.
The MtlR-HPr interaction is conserved within the Enterobacteriaceae family. (A) Amino acid sequence alignment of MtlRs among E. coli and Vibrio species using ClustalX2. The conserved residues are highlighted in black (100%) and gray (75%). The residues involved in the interaction with HPr are indicated by red arrows. Four residues tested in Fig. 3C are indicated by asterisks. ec, E. coli; vp, V. parahaemolyticus; vv, V. vulnificus; vc, V. cholerae. (B) Comparison of the HPr-binding surface on ecMtlR with the corresponding surface on vpMtlR (colored in red). The residues that are not identical between ecMtlR and vpMtlR are labeled with numbers on their surfaces. The conserved hydrophobic pockets are indicated by arrows. (C) Tests of interaction between MtlRs and HPrs. Both E. coli and V. vulnificus proteins (HisMtlR and HPr) were used for the binding tests. His-tagged MtlR (200 μg) from E. coli or V. vulnificus was mixed with HPr (200 μg) from E. coli or V. vulnificus and subjected to Talon metal affinity chromatography. After a three-time wash, bound proteins were eluted and analyzed by 4–20% SDS-PAGE and staining with Coomassie brilliant blue.