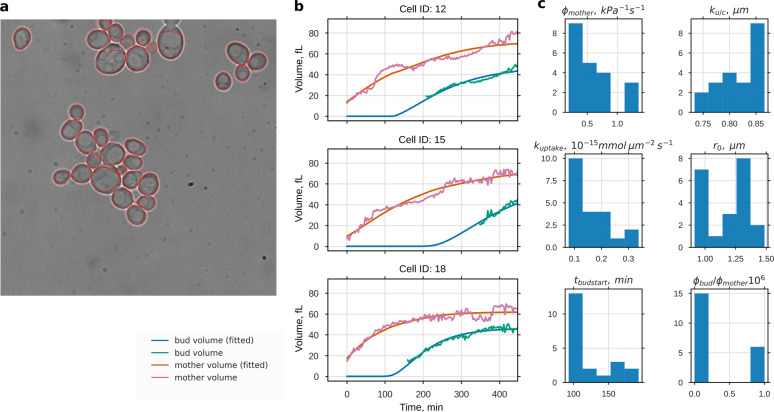
Fig. 4.
Growth parameter estimation from time series of bright field microscopy images. a Cross-sectional areas of 21 single yeast cells were automatically detected from transmission microscopy images and followed over time. Estimated volume increases of mother and bud were used to fit the model parameters: import rate of osmolytes kuptake, import and degradation ratio of osmolytes ku/c, cell wall extensibility of the mother ϕmother, and the extensibility ratio of bud and mother ϕbud/ϕmother. Time of bud start tbudstart and initial osmotic radius r0 were used as free initial conditions. Three measured volume trajectories and corresponding fits are shown in b (for the complete data set see Supplementary Fig. 7). Histograms of the resulting parameter values are shown in c. Except for ϕbud/ϕmother we could assign all of the parameters with low variability, despite the high variability in the volume development in these data