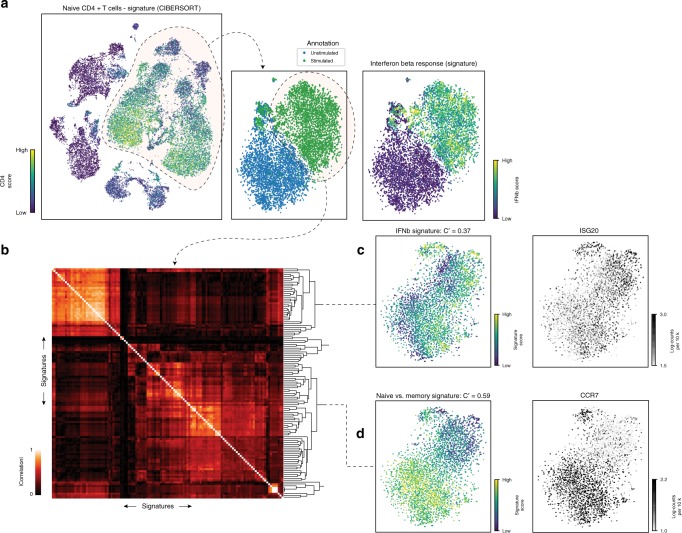
Fig. 2.
Vision highlights within-group and between-group variation within Lupus PBMCs. a Stimulated and unstimulated PBMCs from all eight donors. A cell type signature (from CiberSort52) is used to identify the CD4 T cell cluster. Isolating the CD4 T cells reveals that interferon beta stimulation is the dominant component of variation. This is identified through the use of an interferon-beta stimulation signature and confirmed with the cell annotations. b Further isolating only the stimulated CD4 T cells, signatures with significant local autocorrelation are clustered based on the absolute correlation of their per-cell signature scores to reveal signature modules. c One such module describes the naive vs. memory axis of variation (signature from ref. 34 via MSigDB, and naive T cell marker gene CCR7 shown). d A second module describes variable activation of the interferon response pathway within the stimulated CD4 cells (signature from ref. 33 and interferon-induced gene ISG20 shown)