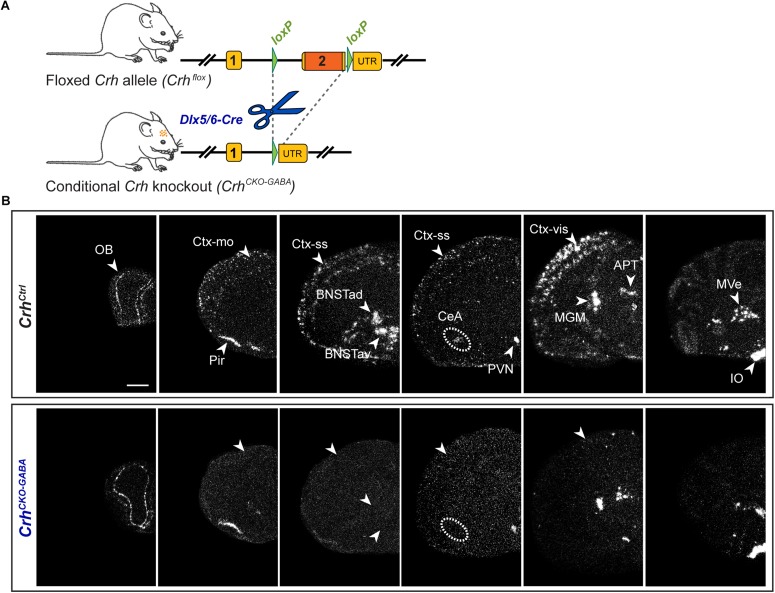
FIGURE 1.
Crh deletion pattern in conditional CrhCKO–GABA mice. (A) Schematic representation of the previously established conditional inactivation of the Crh gene (Dedic et al., 2018a). Exon 2 is flanked by loxP sites. Breeding of Crhflox mice to Dlx5/6-Cre driver mice resulted in Crh deletion specifically in GABAergic forebrain neurons. UTR, untranslated region. (B) Expression of Crh mRNA was assessed by ISH in control and CrhCKO–GABA mice using a riboprobe, which detects the 3′UTR of Crh. Dark-field photomicrographs depict the specific Crh deletion pattern in CrhCKO–GABA mice. Areas of interest are highlighted with arrowheads and dashed lines. Images are representative of three separate experiments. APT, anterior pretectal nucleus; CeA, central amygdala; BNSTad/v, anterior bed nucleus of the stria terminalis dorsal/ventral; CtxII/III, CtxV/VI, cortical layers; IO, inferior olive; MGM, medial geniculate nucleus; MVe, medial vestibular nucleus; Ctx-mo, motor cortex; OB, olfactory bulb; Pir, piriform cortex; PVN, paraventricular nucleus of the hypothalamus; Ctx-ss, somatosensory cortex; Ctx-vis, visual cortex. Scale bar represents 1 mm.