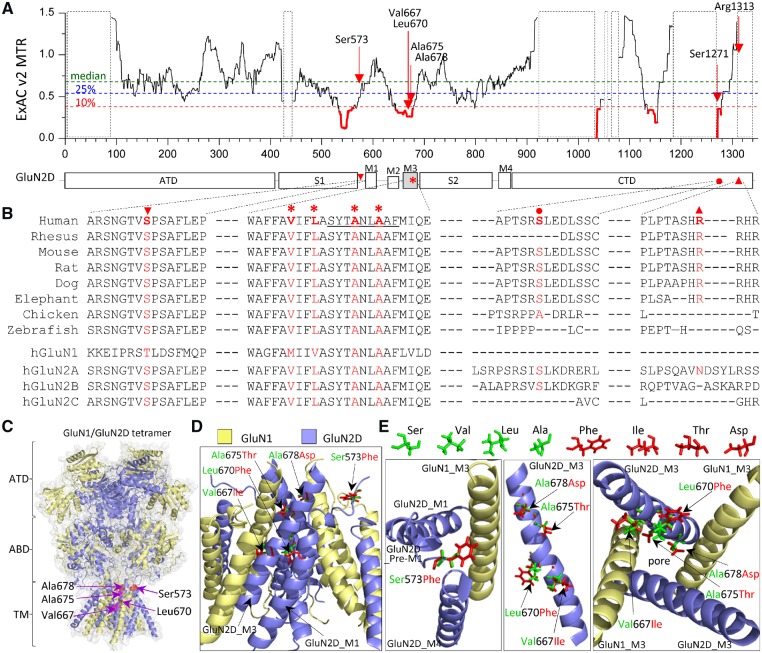
Figure 3.
Location of seven missense variants in GRIN2D/GluN2D. (A) Intolerance analysis of genetic variation across functional domains within GRIN2D/GluN2D. observed/expected mutant ratios (OE-ratio) below the 10th percentile are in red and indicate the regions under purifying selection (Traynelis et al., 2017). Residue Ser573 is located in pre-M1 helix, linker region between agonist binding domain S1 and transmembrane domain M1. Residues Val667, Leu670, Ala675, and Ala678 are located in transmembrane domain M3, one of the least tolerant regions. Residues Ser1271 and Arg1313 are located in intracellular CTD. The regions highlighted with a dashed box have insufficient data available. ATD = amino-terminal domain; S1, S2 comprise the ABD (agonist binding domain); M1, M2, M3, M4 comprise the transmembrane domain; CTD = carboxyl-terminal domain. (B) The residues of Ala675 and Ala678 are highly conserved across other vertebrate species and all other GluN subunits. The residues Ser573, Val667 and Leu670 are highly conserved across other vertebral species and all other GluN2 subunits, but not the GluN1 subunit. The residues Ser1271 and Arg1313 are conserved in most vertebral species evaluated, but not in other GluN subunits. (C) Homology model of the GluN1/GluN2D receptor built (Li et al., 2016) from the GluN1/GluN2B crystallographic data (PDB: 4PE5; Lee and Chung, 2014; Karakas and Furukawa, 2014) is shown as ribbon structure overlaid by space-filled representation. The GluN1 subunit is yellow and the GluN2D subunit is blue. The positions of p.Ser573Phe, Val667Ile, Leu670Phe, Ala675Thr and Ala678Asp are highlighted by red in the pre-M1 helix and M3 domain. (D) The tetrameric GluN1/GluN2D transmembrane region (ATD and ABD removed) viewed from the side. (E) Potential interaction between GluN2D pre-M1, GluN1-M3, and GluN2D-M4 top-down through the pore (left), side view of one GluN2D M3 domain (middle), and M3 domains of two GluN1 and two GluN2D viewed from the bottom through the pore (right). Wild-type residues Ser, Val, Leu and Ala are green, and mutant residues Phe, Ile, Thr, and Asp are shown as red.