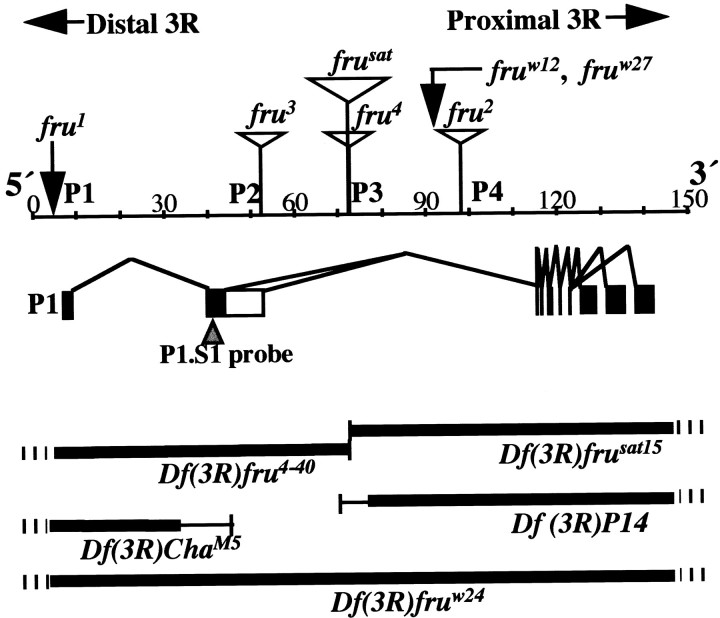
Fig. 1.
The fruitless gene and genetic variations at the locus. This large (∼130 kb) gene contains at least four promoters (Ps); the 5′-most promoter (P1,left) controls the production of primary transcripts that are sex-specifically spliced with respect to the second exon (Ryner et al., 1996; Goodwin et al., 2000). P1-promoted RNA species are diagrammed in the middle panel. The principal alternative splicings of interest occur near the 5′ (left) end. (There are additional such splicings near the 3′ end, designated by three black rectangles on theright that result in different kinds of Zn-finger pairs near the C termini of FRU protein isoforms.) The 5′ sex-specific splicings result in a male mRNA of which relatively 5′ coding sequences (5′ ORF) are translated to produce 101 male-specific amino acids bound to the remainder of the residues that are encoded by relatively 3′ sequences emanating from the right part of the gene. This protein is called FRUM, which is specifically detected by an antibody generated against the male-specific N-terminal residues (Lee et al., 2000). In females, the 5′ ORF runs into a stop codon after nucleotides encoding 94 amino acids (because of the alternative splicing referred to above). A ∼280-nucleotide probe applied in this study to detect sex-specifically spliced transcripts (cf. Lee et al., 2000) corresponds to sequences in the second exon, designated by a black rectangle (middle left) and pointed to by an inverted triangle. Sex-nonspecific promoters (P2, P3, andP4) are used to generate transcripts that lack the male-specific 101 residues and are believed to be associated with vital fru functions that operate in both sexes (Ryner et al., 1996; Anand et al, 2001). Such functions are inferred (in part) from the effects of fruw12 andfruw27, which are translocation and inversion breakpoints that cause late-developmental lethality when they are homozygous or heterozygous for a deletion that eliminates the entire locus. fruw24 is such a deletion (Df), indicated by the thick black line (this and other such lines designate deleted material); hash marks (for this and the otherDfs) indicate that the deletion extends well beyond the locus. The four additional Dfs that were applied have breakpoints within the locus (Ito et al., 1996; Ryner et al., 1996;Anand et al, 2001), as indicated by thin vertical lines(the thin horizontal portions of these Dfindicators imply not-quite-certain breakpoint determinations). Homozygous-viable fruitless mutants are caused, in one case, by an inversion breakpoint (fru1) and, in the remaining cases, by transposon inserts (open triangles) inserted at intragenic locations as determined by Ito et al. (1996), Ryner et al., (1996), and Goodwin et al. (2000). These and other features of the diagram are based on results in these three reports, as well as information obtained by interrogating theDrosophila genome database at www.fruitfly.org with sequences of various fru cDNAs (Lee et al., 2000) and molecular determination of the fru1inversion breakpoint, which was found to be 3.3 kb upstream (to theleft) of the transcription start site for RNAs generated under the control of the P1 promoter. The beginning of a 7 bp consensus-sequence for the latter starts 28 bp downstream of the transcription start (T. Carlo, S. F. Goodwin, J.-C. Billeter, L. C. Ryner, B. S. Baker, and J. C. Hall, unpublished observations).