Fig. 1.
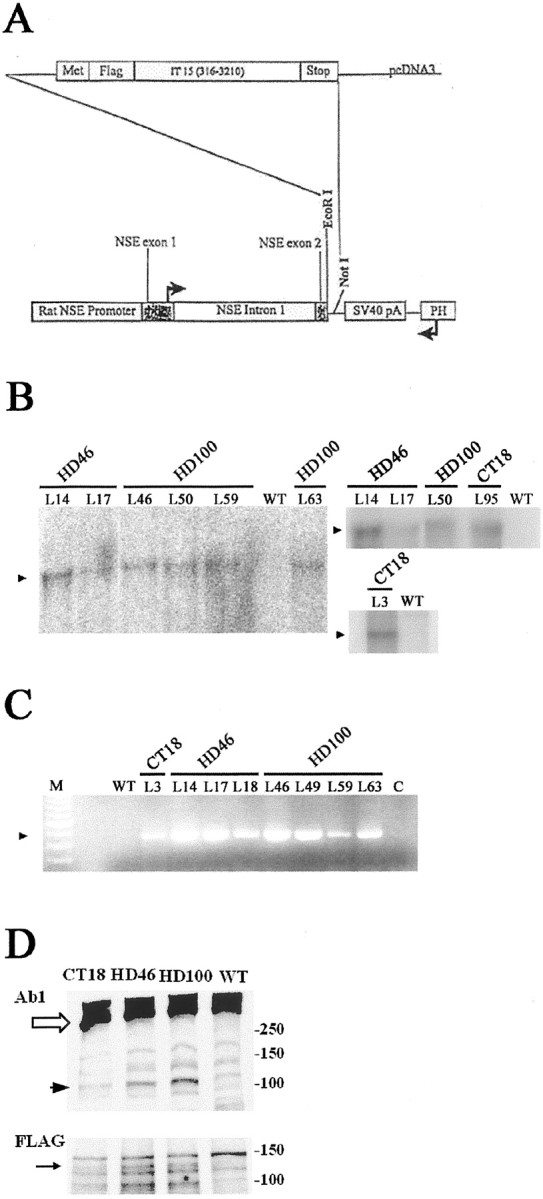
Design and expression analysis of transgenes CT18, HD46, and HD100. A, Map of construct used for transgene vector. Bases 316–3210 of the human huntingtin cDNA sequence (IT15), modified to contain 18, 46, or 100 CAG repeats, were cloned into AcMNPV transfer plasmid NSE4-BV. A rat neuron-specific enolase promoter regulates expression. An N-terminal epitope tag, FLAG, was included to permit differential identification of the transgene-encoded huntingtin product. Northern blot and RT-PCR analyses confirm RNA expression from CT18, HD46, and HD100 transgenes.B, The Northern blot demonstrates transgene expression in different HD46 and HD100 lineages (denoted above thelanes). B, Left andTop right, Total RNA (10 μg) per lane.B, Bottom right, Total RNA (30 μg) per lane. An arrowhead (▴) marks the bands corresponding to transgene-encoded RNA. Wild-type controllanes show no transgene-derived message.C, RT-PCR results from CT18, HD46, and HD100 lineages. Lane M, A 100 base pair ladder; lane C, no RNA control. Anarrowhead (▴) marks the position of the expected RT-PCR product. Expression is seen in all transgenic lineages but is absent from the transgene-negative (WT) control.D, Western blot analysis of huntingtin (top) and FLAG (bottom) immunoreactivity in WT, CT18, HD46, and HD100 mice. D,Top, A product of the expected size (∼120 kDa,black arrowhead) is present in the transgenic mice, but not in WT. Native huntingtin is denoted with a white arrow. The signal intensity for the ∼120 kDa product is stronger in the HD mice compared with CT. Huntingtin is detected with antiserum Ab1. D, Bottom, A product of the expected size (∼120 kDa, black arrow) is present in the transgenic mice, but not in WT.
