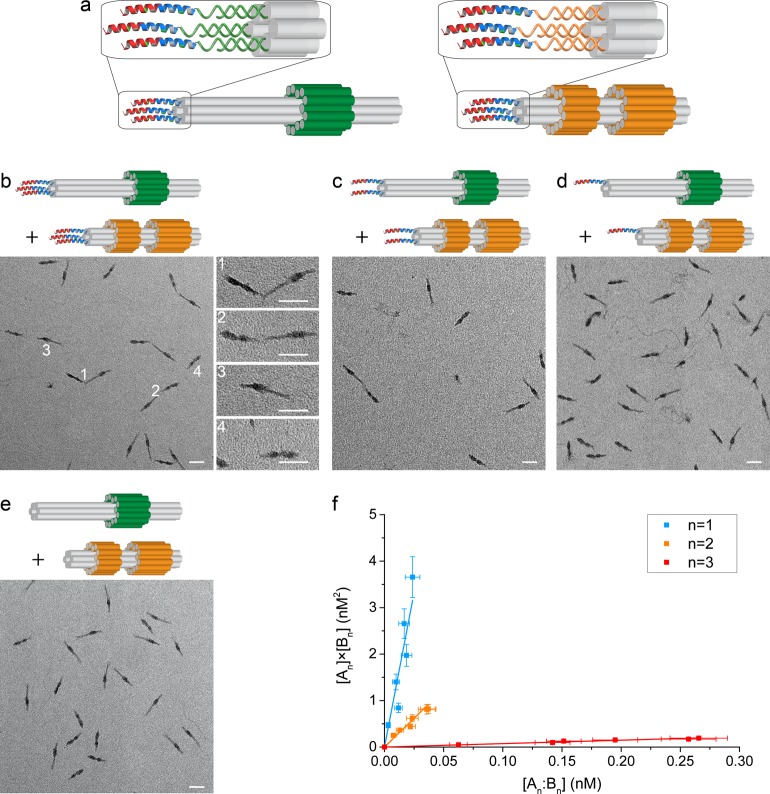
Figure 3.
Directed assembly of DNA nanostructures through interactions of peptide–oligonucleotide conjugates. (a) DNA origamis A3 and B3 decorated with three peptides. (b–e) Representative TEM images of the assembly of peptide-decorated DNA origamis. The distribution between species depends on the number of peptides, n, displayed on each origami: (b) n = 3, (c) n = 2, (d) n = 1, and (e) n = 0. White numbers in (b) indicate representative heterodimers (1 and 2) and monomers (3 and 4). The initial concentrations of assemblies An and Bn were, respectively, 0.61 and 0.84 nM (b, n = 3), 0.84 and 1.09 nM (c, n = 2), 1.69 and 2.39 nM (d, n = 1), and 0.68 and 0.74 nM (e, n = 0). Scale bars: 50 nm. (f) Dissociation constants of heterodimers of origamis functionalized with different numbers of peptides were determined from linear fits (solid lines) to plots of [An] × [Bn] as a function of [An:Bn] over a range of total monomer concentrations from 0.25 to 2.4 nM. Data were weighted by estimated uncertainties (SI Tables S4, S5 and supporting discussion).