Abstract
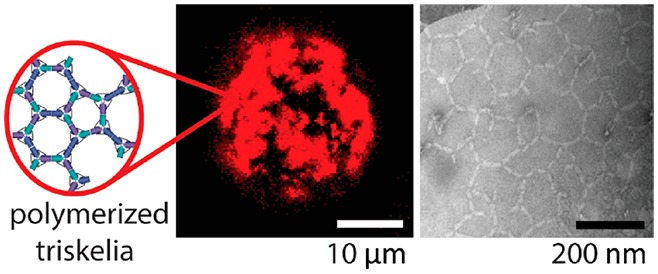
We describe the triggered assembly of a bioinspired DNA origami meshwork on a lipid membrane. DNA triskelia, three-armed DNA origami nanostructures inspired by the membrane-modifying protein clathrin, are bound to lipid mono- and bilayers using cholesterol anchors. Polymerization of triskelia, triggered by the addition of DNA staples, links triskelion arms to form a mesh. Using transmission electron microscopy, we observe nanoscale local deformation of a lipid monolayer induced by triskelion polymerization that is reminiscent of the formation of clathrin-coated pits. We also show that the polymerization of triskelia bound to lipid bilayers modifies interactions between them, inhibiting the formation of a synapse between giant unilamellar vesicles and a supported lipid bilayer.
Keywords: nanotechnology, clathrin-inspired nanostructure, DNA origami, membrane interaction, biomimetic
Much of our understanding of membrane biology arises from direct observation of active cellular membrane processes, including the dynamic modulation of the size, shape, and properties of the membrane by the action of membrane-associated proteins.1 Proteins such as annexins and s-layer proteins assemble into dense networks to impart structural rigidity and protection from the environment.2,3 Peripheral membrane proteins such as clathrin, dynamin, SNARE proteins, and BAR-domain proteins are associated with site-specific membrane curvature.4−6 Clathrin, which has a characteristic triskelion shape, is a well-studied example.4 During endocytosis, clathrin self-assembles to form a polyhedral lattice that gradually coats the membrane. Upon polymerization of the protein, the lipid bilayer invaginates and eventually detaches from the cell membrane (assisted by the action of dynamin)6 to diffuse into the cytoplasm.
DNA nanotechnology provides the opportunity to construct simplified biomimetic models for the study of biological systems.7−13 DNA can bind to a lipid membrane electrostatically and through hydrophobic interactions of unpaired bases.14,15 Conjugation with lipophilic moieties such as cholesterol improves the ability of DNA origami nanostructures to associate with, and diffuse on, lipid bilayers,7,8,14,16−19 allowing the assembly of ordered arrays.7,8,11,14,20 Recently, Xu etal. demonstrated DNA structures used as a support for the organization of SNARE proteins leading to membrane fusion.12 DNA origamis can also be designed to mimic the shapes and functions of membrane-modifying proteins: Franquelim etal. demonstrated an origami structure for membrane binding and sculpting,11 and Grome etal. explored a spiral DNA origami, inspired by dynamin, that can polymerize to induce membrane tubulation.13
In this paper, we study the properties and interactions of a DNA origami triskelion, which models the structural properties of clathrin and can be programmed to polymerize into flexible, extended, membrane-bound assemblies. We observe nanoscale membrane deformations, which resemble the effects of clathrin assembly, when DNA origami triskelia are polymerized on a lipid monolayer coating an EM grid. We also show that polymerization of the origami triskelia affects the interactions between lipid bilayers.
Results and Discussion
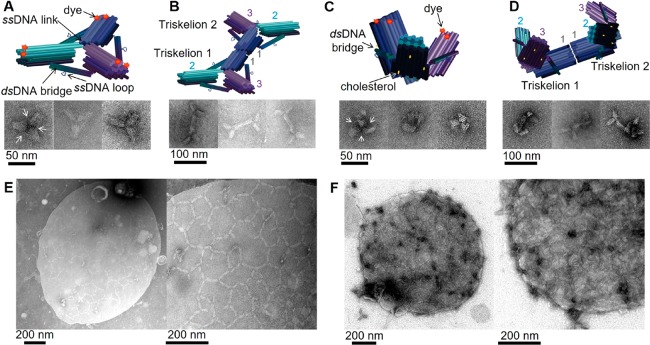
We designed a three-arm DNA origami nanostructure whose shape resembles that of the clathrin triskelion (Figure 1, Supporting Figure S1).21 The DNA origami is approximately 20 times more massive than its natural counterpart.22 Each triskelion arm consists of 28 parallel DNA helices organized on a honeycomb lattice to create a 13 nm diameter bundle that is 30 nm long. The angles between the arms of the DNA triskelion are constrained by rigid three-helix bundles that form bridges connecting the midpoints of each arm (Figure 1A, Supporting Figure S1). We sought to control the shape of the triskelia by using bridges of different lengths: the flat triskelion has arms of 92 base pairs (bp) (angle between arms approximately 120°, Figure 1A,B), and the curved triskelion has arms of 84 bp (forming a triangular pyramid with a height of approximately 18 nm, Figure 1C,D). Arms are linked to each other, where they meet near the center of the structure by single-stranded DNA links, formed by routing the scaffold strand between arms, and by the bridges between arms (through which the scaffold also runs). Triskelia can be programmed to assemble into extended arrays on addition of staples that link arms end-to-end. An edge in this network is approximately 60 nm long, twice the dimension of the natural clathrin lattice.23
Figure 1.
Triskelion monomers, dimers, and arrays. (A–D) Designed structure and transmission electron microscopy (TEM) micrographs of (A) flat triskelion monomer; (B) dimer formed by linking arms 1 end-to-end; (C) curved triskelion monomer; and (D) curved dimer. Cylinders represent DNA helices. White arrows point to bridges, visible on some of the electron micrographs. (E) TEM micrograph of lipid monolayer to which flat triskelion dimers were attached before their polymerization was triggered by addition of DNA polymerization staples with 6 nt sticky ends linking arms 2 and 3. A discrete, approximately circular membrane structure is visible, covered by a partially ordered triskelion array with hexagonal and pentagonal cells. (F) as in E but using curved triskelion dimers. Magnified images in the right-hand panels of E and F are of structures similar to those shown on the left. We attribute the formation of discrete, triskelion-coated membrane structures (E and F) to local deformation (budding) of the membrane induced by the nanostructure.
We functionalized the triskelion with three cholesterol groups on the broader “bottom” surface of each arm to enable direct attachment to lipid membranes. Six Alexa647 fluorophores are attached to the narrower upper layers of DNA helices (Supporting Figure S1) for visualization by fluorescence microscopy. In the right-hand electron micrograph in Figure 1C the honeycomb cross-section of the bundles of helices in the origami arms is clearly resolved: the “top” of each arm, a layer of four DNA helices, is at the center of the 3-fold structure, indicating that the triskelion has been forced into a conformation in which the cholesterol anchors lie on the convex side of the distorted structure, as shown in the diagram (Supporting Figure S2). Other images indicate that triskelia also fold with the cholesterols on the inner side (Supporting Figure S3).
Clathrin self-assembles into arrays as a result of weak attractive interactions distributed along its arms.22,24 For design purposes it was easier to localize the linking sites of our artificial triskelion at the extremities of its arms, doubling the distance between two attached origami centers compared to that of clathrin. Individual DNA triskelia can be linked through the addition of DNA staples that bind to the origami scaffold at the ends of the arms. Most of the experiments described used preformed triskelion dimers, which we found to give better-formed arrays when bound to a membrane and polymerized. Dimers are formed during origami assembly using six dimerization staples, each of which binds to scaffold domains at the ends of both of the arms 1 of the two component triskelia, forming six parallel connections between them (Figure 1B,D and Supporting Figures S4 and S5). Dimers can be linked into arrays by adding two sets of six DNA polymerization staples (12 strands in total), each of which hybridizes to the scaffold at the ends of one of the free arms (arms 2 and 3), creating 10 overhanging 6-nucleotide “sticky ends” at the end of each arm (Supporting Figures S4 and S6). These sticky ends are designed such that hybridization of the sticky ends displayed on arm 2 of one dimer to those on arm 3 of another links the two arms together: this connectivity is consistent both with a hexagonal array and with the formation of pentagonal cells (Supporting Figure S4D), allowing curvature. The pattern of connections between pairs of arms is such that the two origamis are aligned with the membrane-binding faces orientated in the same direction.
Triskelion networks were observed by transmission electron microscopy (TEM). Networks formed by polymerization in solution, in the absence of lipids, are generally extended and poorly ordered; occasional polygons are observed (Supporting Figure S7). Triskelia inserted into the membranes of small unilamellar vesicles (SUVs) formed dense coatings around the vesicles (Supporting Figure S8). However, distortion of the vesicles by the relatively harsh staining and drying protocol required for TEM (Materials and Methods) precluded clear identification of the effect of the DNA origami on membrane shape.7
Polymerization of triskelion dimers on supported lipid monolayers was imaged by TEM using an apparatus adapted from that developed in the group of McMahon (Supporting Figures S9, S10 and Materials and Methods).25,26 Addition of a small excess of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, 10–20% more than monolayer coverage) to a pool of buffer in a Teflon well leads to the formation of a lipid monolayer, which can be transferred to a gold TEM grid, stained with uranyl acetate, and observed by TEM (Supporting Figure S11). Flat triskelion dimers injected into the well attach readily to the lipid surface that covers the grid and polymerize into an extended network reminiscent of clathrin assembly (Figure 1E). Curved triskelia form denser, less regular, networks (Figure 1F). In both cases distinct, isolated, clusters, approximately circular in projection, are observed after, but not before, triskelion polymerization (Figure 1E and F, Supporting Figures S11–S13). These structures are consistent with local deformation (budding) of the monolayers induced by the formation of triskelion arrays and are similar to TEM images of clathrin-coated pits on lipid monolayers.25−27 In the case of flat triskelia, the circular clusters are frequently partly circumscribed by high-contrast crescent-shaped regions, characteristic of the projection image of a partially collapsed bleb.28 This is particularly clear in Supporting Figure S11E, in which two such structures are superimposed. In the case of curved triskelia, the clusters are more densely stained with little evidence of collapse. Observation of on-edge triskelia at the edges of the circular structures supports our interpretation that the membrane is deformed (Supporting Figure S13).
The membrane blebs induced by the two triskelion variants are qualitatively similar, despite that fact that one triskelion is designed without intrinsic curvature and the surface of the other is much more curved than the blebs themselves. In neither case can our triskelia be forcing the membrane to conform to an intrinsic array curvature. This is consistent with the observation that the clathrin protein can form planar as well as curved arrays, yet clathrin alone (if bound to the membrane) is sufficient to induce the formation of spherical buds.29 It has been suggested that natural membrane bending, including the formation of clathrin-coated pits, is not directly dependent on the details of protein structure but is driven by crowding of membrane-anchored proteins:30 protein crowding alone is even sufficient to drive membrane fission.31 We suggest that a similar mechanism drives the membrane deformation that we observe: triskelion polymerization induces locally dense membrane coverage and (in contrast to better-ordered and more rigidly connected arrays of membrane-bound DNA nanostructures)8 the disordered triskelion lattice provides an entropic drive for the membrane to curve away to relieve crowding.
The induction of membrane curvature through polymerization of DNA triskelia is qualitatively different from the effects of membrane-bound DNA nanostructures reported previously. Most published studies of DNA nanostructure arrays on membranes are of planar arrays on planar membranes. Indeed, tightly packed and well-ordered DNA arrays have been shown to induce planar deformations of naturally curved GUV membranes.8 Where increases in membrane curvature have been achieved by membrane-bound DNA nanostructures, it has been through strong interactions that force the membrane to conform to the intrinsic curvature of the nanostructure.9−11,13 It has been suggested that the natural role of the similarly shaped proteins that inspired these nanostructures is to sense, rather than to induce, membrane deformation.32−34
In order to study of the dynamics of triskelion–membrane interactions, we used fluorescence microscopy to observe triskelion assembly on lipid bilayers, using both giant unilamellar vesicles (GUVs) and supported lipid bilayers (SLBs). These experiments do not resolve the sub-micrometer membrane deformations described above but do enable investigation of the effects of triskelia on membrane interactions. The two triskelion types (flat and curved) behaved similarly in these experiments.
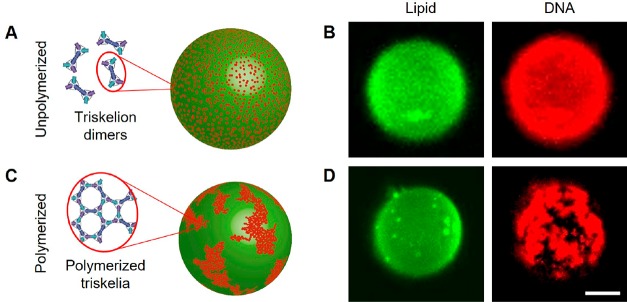
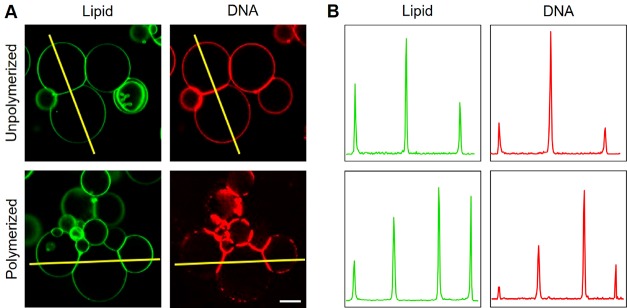
GUVs, comprising DOPC with 0.1 mol % fluorescently labeled 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (Atto488-DOPE), were prepared by electroformation in 52 mM sucrose.16,17 Triskelion dimers with Alexa647 labels (Supporting Figure S1) were assembled in equiosmolar TE-MgCl2 buffer, incubated for at least 10 min with the GUV suspension diluted 20-fold in the same TE-MgCl2 buffer (Supporting Figure S14), and observed using confocal microscopy. Dimers were observed to bind to membranes homogeneously and diffuse freely (Figure 2A,B and Supporting Movie M1). When polymerization of the dimers was triggered by addition of polymerization staples, DNA triskelia assembled into dense networks on the GUVs (Figure 2C,D, Supporting Movies M1–M4), observable as discrete, diffusing aggregates. We do not see evidence of planar deformations similar to those induced by the densely packed nanostructure arrays reported by Czogalla etal.8 Membrane-bound DNA triskelion dimers, before and after polymerization, are enriched at GUV–GUV interfaces (Figure 3).
Figure 2.
DNA triskelia interacting with giant unilamellar vesicles. (A, C) Inferred distributions of DNA triskelion dimers on GUVs. Unpolymerized DNA triskelia are homogeneously distributed on the GUV surface; polymerization, triggered by addition of polymerization staples, causes triskelia to assemble into arrays in mesoscopic domains. (B, D) Confocal micrographs corresponding to a 500 nm thick section through the top of the GUV. The formation of large clusters of curved triskelia on polymerization is evident but has no significant effect on the lipid distribution. Scale bar: 10 μm.
Figure 3.
Confocal micrographs of GUV–GUV interfaces. (A) DNA triskelia (curved) are enriched at interfaces between GUVs both before and after the addition of polymerization staples. Scale bar: 20 μm. (B) Intensity profiles along the lines indicated in A show the accumulation of both unpolymerized and polymerized triskelia at GUV–GUV interfaces.
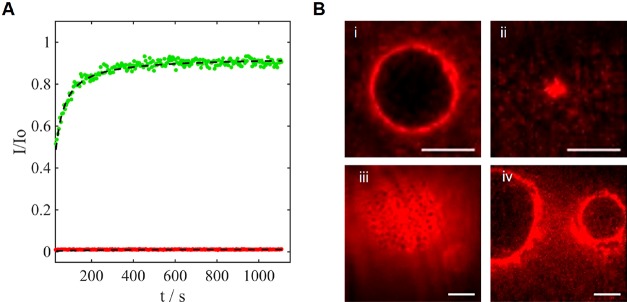
We performed similar experiments to examine the assembly of fluorescently labeled triskelia on SLBs using epifluorescence and total internal reflection fluorescence (TIRF) microscopy. In contrast to their behavior on GUVs, dimer diffusion and triskelion polymerization on the SLB were inhibited: fluorescence recovery after photobleaching (FRAP) confirmed the presence of mobile lipids but immobile triskelion dimers (Figure 4A). GUV–SLB interfaces were formed by sedimentation of sucrose-containing GUVs on an SLB washed in less-dense TE-MgCl2 buffer. Triskelia diffusing freely on the GUV were excluded from this interface and accumulated at its edge (Figure 4B i; see also Supporting Figure S15 i). Triskelion dimers added after GUV–SLB interface formation also accumulated at the junction between the two bilayers (Figure 4B iv and Supporting Figure S15 iv). In each of these experiments, the initial contact between a GUV and an SLB expands to form a planar, circular, interface within seconds. However, when the triskelia bound to GUVs are polymerized by addition of polymerization staples before the two bilayers were brought into contact, formation of the contact interface is inhibited for at least several minutes (Figure 4B ii, Supporting Figures S15 ii and S16), When triskelion dimers are initially bound to the SLB rather than to the GUV, triskelia are partially excluded from the interface (Figure 4B iii and Supporting Figure S15 iii). The difference in behavior between triskelia initially bound to the GUV (unpolymerized triskelia escape the interface) and to the SLB (slowly diffusing triskelia remain at the interface) confirms that the interactions between triskelia and the lipid bilayers are directional, consistent with stable insertion of cholesterol into the first membrane encountered.
Figure 4.
DNA triskelia on supported lipid bilayers and at GUV–SLB interfaces. (A) Fluorescence recovery after photobleaching (FRAP) analysis of fluorescently labeled curved DNA triskelia (Alexa647, red) and lipid (Atto488, green) shows that the lipids in the SLB are mobile, whereas the DNA triskelia are immobile. (B) TIRF micrographs of fluorescently labeled triskelia at GUV–SLB interfaces: (i) triskelia added to GUV before synapse formation are excluded from the interface; (ii) triskelia added to GUV and polymerized before synapse formation inhibit interface formation; (iii) triskelia added to SLB before synapse formation remain at the interface; (iv) triskelia added after synapse formation are excluded from the interface. Triskelia used (see Supporting Figure S15 for the other type): curved (i, iii); flat (ii, (iv). Scale bars: 10 μm.
Accumulation of triskelia at the interfaces between GUVs is consistent with passive diffusion-mediated trapping. It implies that triskelia mediate an attractive interaction between the membranes by binding to both, an important mechanism in cell adhesion;35,36 it is consistent with the observation that DNA origamis can bind to lipid membranes even in the absence of cholesterol anchors.37 In contrast, the exclusion of triskelia from an SLB–GUV interface (where GUV-bound triskelia are able to diffuse away from the interface) and the inhibition of interface formation by polymerized triskelia (whose escape is hindered) suggest that there is a repulsive interaction between GUV and SLB mediated by triskelia. The marked asymmetry between triskelion-mediated GUV–GUV and GUV–SLB interactions is intriguing. All membranes have the same lipid composition: the most obvious difference between them is that the SLB is planar and closely bound to a glass surface, whereas the GUV bilayer is constrained only by its natural elasticity and any residual difference in osmotic pressure across it. We hypothesize that the stable incorporation of a triskelion array at a GUV–GUV interface requires significant distortion of the membranes to conform to the far-from-planar triskelia: the SLB is incapable of this distortion.
Conclusion
We have demonstrated the controlled formation of extended DNA origami triskelion arrays on lipid bilayers by electron and optical microscopy. We have observed that polymerized networks of triskelia can induce sub-micrometer deformation of a lipid monolayer, which is reminiscent of the formation of clathrin-coated pits. Triskelia also modulate lipid interfaces: they mediate an attractive interaction between free bilayers but can inhibit the formation of an interface when one bilayer is bound to a rigid substrate. These results help demonstrate the potential of biomimetic membrane-associated nanostructures as tools to control the dynamic behavior of lipid membranes and their shapes and interactions. We anticipate that the exploration of the design of membrane-modifying nanostructures will lead both to a greater understanding of natural processes and to biomimetic systems for signaling, synthesis, and reproduction based on membrane-bound compartments.
Materials and Methods
1,2-Dioleoyl-sn-glycero-3-phosphocholine was purchased from Avanti Polar Lipids (Alabaster, AL, USA). 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine labeled with Atto488 (Atto488-DOPE) was purchased from AttoTec (Germany). All chemicals and solvents were of analytical grade, purchased from Thermo Fisher Scientific or Sigma-Aldrich, and used without further purification. All DNA strands, including Alexa647- and CholTEC- conjugates, were provided by Integrated DNA Technologies and used as received. Single-stranded DNA scaffold M13mp18 (p7249) was supplied by VWR International Ltd. (1 μg/μL, in 10 mM Tris (pH 7.5), 1 mM EDTA) and used as received.
Triskelion Synthesis and Purification
The triskelia and triskelion dimers were folded in TE-MgCl2 buffer by thermal annealing ramp on a programmable, temperature-controlled Peltier heat block (Eppendorf Mastercycler thermal cycler) with the following schedule: 65 °C for 15 min; decrease to 58 °C at −1 °C every 5 min; decrease to 40 °C at −1 °C every 60 min; decrease to 20 °C at −1 °C every 5 min.
After a solution of the DNA scaffold and staples was annealed to yield the desired origami, the sample was purified with a hand-packed size-exclusion column. This type of size-exclusion method foregoes centrifugation and relies on gravity to elute the buffer through the resin. The resin used was Sephacryl S-300 (GE Healthcare Life Science) resuspended in folding buffer (1× TE + 14 mM MgCl2). A bed volume of 8 mL is needed to separate the origami from excess staples. Disposable columns (GE Healthcare Life Science, PD-10) were hand-packed with a bottom frit circle followed by the resin stock in appropriate buffer (typically 1× TE + 14 mM MgCl2). The column was rinsed with buffer of twice the bed volume to sediment and equilibrate the column before placing an upper frit circle that protects the resin from drying out. A sample from 50 to 800 μL was then applied to the top of the frit. To reduce dilution, it is important to wait until the sample has been totally absorbed in the resin, and the first fraction was collected before adding more buffer to the reservoir. Typically, 19 fractions of approximately 250 μL are collected per sample and analyzed by AGE (see protocol below). A typical 50 μL sample of 20 nM triskelia usually yields one 250 μL fraction clear of staples at a concentration of 2–3 nM. This method works for triskelion monomer and dimers, as their migration in the resin is similar.
After collection of the fractions from the size-exclusion column, 10 μL of each fraction was loaded with loading buffer (0.25% w/v bromophenol blue (Sigma-Aldrich), 30% w/v glycerol (Sigma-Aldrich) in a well of a 1.5% agarose gel with 0.5× TBE buffer (45 mM Tris base, 45 mM boric acid, 1 mM EDTA) plus 14 mM MgCl2. A potential of 50–60 V was applied along the length of the gel. For long electrophoresis runs, the buffer was replaced halfway in order to maintain a current of approximately 12 mA. The agarose gels were stained with SYBR gold (Thermo Fisher Scientific) for analysis by fluorescence. Fractions found to contain triskelia (typically fractions 10 and 11) were combined and used for confocal experiments. For quality control, the bands of interest localized in the gel were cut out with a scalpel, finely chopped, and pressed between two Parafilm-protected microscope slides to extract the solution. The recovered volume was imaged by TEM.
When triskelia were prepared for experiments that did not require confocal imaging, the triskelia were purified by AGE, stained with SYBR gold, and extracted by cutting the bands out of the gel as described above.
Insertion of Triskelia into Lipid Layers and Polymerization
Unless otherwise stated, polymerization of triskelion dimers and monomers was performed as follows.
Triskelia without Lipid
Purified triskelia (50–100 μL, 0.5–3 nM) in 0.5× TBE buffer plus 14 mM MgCl2 were mixed with 10 equiv of polymerization staples (initial concentration of added solution 2500 nM in 1× TE plus 14 mM MgCl2) and left at room temperature overnight. The sample was then stained with UAc for TEM imaging as described below.
Cholesterol-Bearing Triskelia and SUVs
Purified cholesterol-bearing triskelia (75 μL, 3 nM) in 0.5× TBE buffer plus 14 mM MgCl2 were mixed with 15 μL of extruded SUVs (ca. 0.8 mg mL–1) in 1× TE buffer plus 14 mM MgCl2 and left at room temperature for a minimum of 10 min to allow the cholesterol to insert into the bilayers. Polymerization staples (10× equivalent, 2500 nM initial concentration in 1× TE plus 14 mM MgCl2) were then added, and the sample was left to polymerize at room temperature overnight before TEM imaging.
Cholesterol-Bearing Triskelia and GUVs
Purified cholesterol-bearing triskelia dimers in 1× TE buffer plus 14 mM MgCl2 (20 μL, 1.3 nM) were added to 5 μL of GUV suspension diluted 20-fold in the same TE-MgCl2 buffer (final lipid concentration ∼0.001 mg mL–1) in 52 mM sucrose and left for a minimum of 10 min to allow the cholesterol to insert into the bilayers. For polymerization experiments, polymerization staples (10× equivalent, 2500 nM initial concentration in 1× TE plus 14 mM MgCl2) were added and left to polymerize at room temperature for a minimum of 20 min before imaging.
The protocol for insertion and polymerization of the triskelia for the monolayer assay is described below (see monolayer assay).
TEM Imaging
TEM micrographs were acquired using a FEI Tecnai 12 with an accelerating voltage of 100 kV (Dunn School of Pathology, University of Oxford, UK). The procedure used for grid preparation (Formvar film and carbon coating of copper and gold grids) is described elsewhere.38 A general protocol for sample preparation for imaging is as follows. A 5–10 μL droplet of DNA sample (1–50 nM) was placed on a glow-discharged grid for 3 min with the grid held in a pair of reverse-action tweezers. Excess liquid was removed by carefully dabbing the edge of the grid with a piece of filter paper. The sample was then quickly washed with 7 μL of stain solution (1–2% UAc) and then blotted once more with filter paper. A second droplet of stain was then applied to the adsorbed sample and left on the grid for 10 s before being blotted away.
Lipid Monolayer Assay
A custom-made Teflon block (see Supporting Figures S9 and S10) was placed in a humid chamber (a small sealed box containing a water-soaked tissue on which the Teflon block is placed), and the wells were filled with 40–50 μL of TE-MgCl2 buffer. To form the lipid layer, 1 μL of lipid mixture (DOPC dissolved in hexane 0.025 mg/mL) was carefully placed on top of the buffer in a well. As a negative control, we used 1 μL of pure hexane without lipid (see Supporting Figure S11 A). The block was incubated in the humid chamber for 30 min to allow the hexane to evaporate, leaving a monolayer of lipid on the surface of the buffer. A carbon-coated gold EM grid was placed, carbon side down, onto the top of each buffer droplet. Grids were not glow discharged before use, as a hydrophobic carbon film is required to adhere to the hydrophobic lipid tails of the monolayer. Cholesterol-bearing triskelia were injected into the well through a side channel, under the EM grid (final concentration of 0.1–5 nM), and left for 30 min. The wells were then washed with fresh buffer to remove noninserted triskelia from the bulk solution. If the triskelia were to be polymerized, we then injected the polymerization staples and left the sample between 6 and 12 h in the humid chamber before removing the grid. The grid was removed by injecting approximately 20 μL of buffer into the side injection port to raise the grid up above the surface of the Teflon block. The grid was removed with forceps and lifted vertically off the droplet. The gold grid was stained as described above without delay to prevent uncontrolled drying of the surface.
Fluorescence Microscopy
GUVs and SLBs were imaged using a combination of confocal, epi-fluorescence, and TIRF microscopies. Epi-fluorescence and TIRF microscopy were performed with a 60× TIRF oil-immersion objective on an inverted microscope (Nikon Eclipse Ti-E). Continuous-wave laser illumination at 474 nm (∼50 μW) and 644 nm (∼120 μW) (Vortran Laser Technology Inc., CA, USA) was used to excite fluorescence for both TIRF and epi-fluorescence modes. The resultant fluorescent signals were transmitted through a ZT488/640rpc dichroic (Chroma Technology Corp) before being separated by an Optosplit II module (Cairn Research) with an FF660-DiO2 dichroic and band-pass filters (Semrock) (680/42 and LP-664 on the red channel and 550/88 on the blue channel). Samples were imaged on an electron-multiplying CCD camera (Andor iXon+ 860) at 50 Hz. For FRAP experiments 0.5 mol % Atto488-DOPE was added to DOPC prior to SUV generation and SLB formation.
Confocal microscopy was carried out in the Nikon Imaging Centre, King’s College London. Imaging was performed with 20× and 40× objectives, and 488 and 642 nm diode lasers were used to illuminate the samples (50 μW).
SUV Formation
SUVs (liposomes) were formed by rehydrating a lipid film. A DOPC lipid stock solution (100 μL, 10 mg/mL in chloroform) was added to a glass vial and dried under vacuum overnight (or for a minimum of 2 h) to remove traces of solvent. The dried lipid film was then rehydrated in 1 mL of buffer and left at room temperature for 1 h without stirring. This facilitates the detachment of the film from the bottom of the vial and forms a suspension of polydisperse multilamellar vesicles. The glass vial was then vortexed to produce a homogeneous turbid solution. The turbid lipid solution was either sonicated with a diving tip or subjected to cycles of freeze–thaw to break the large multilamellar vesicles into smaller liposomes. If sonicated, the glass vial was placed in a beaker of cold water and sonicated (0.5 s every second for 5 × 60 s, 80% power, probe tip sonicator (Vibra Cell, 3 mm titanium probe). The sonication tip was then washed by starting a 1 min cycle of sonication in a mixture of 1:1 EtOH–isopropanol followed by a 1 min cycle in water. If the vial was subjected to freeze–thaw cycles, the vial was first frozen by plunging it into liquid nitrogen, followed by a hot water bath (approximately 40 °C). The freeze–thaw cycle was repeated five times or until the turbidity of the solution was reduced. Lipid aggregates and larger vesicles were removed by extrusion through a 0.2 μm polycarbonate membrane (Whatman) held within a mini-extruder (Avanti Polar Lipids) 21 times. Liposomes were stored at 4 °C under an argon atmosphere and used within 1 week. When mixed with triskelia and imaged by TEM, the SUV solution was diluted 10-fold. Electron micrographs were considered only if the large majority of the grid had homogeneous staining and coverage, so that the images taken are representative of the entire grid.
Lipid Bilayer Formation
SLBs were prepared on glass coverslips by vesicle fusion.39 Lipid mixtures in chloroform (40 μL of 25 mg mL–1 DOPC) were dried under nitrogen and placed under vacuum overnight. The dried lipids were hydrated in 2 mL of water and vortexed before tip sonication (15 min). The resulting clear suspension of SUVs was centrifuged (3 min; 14000g) before the supernatant was separated and retained.
Glass coverslips (VWR, Menzel Gläser #1) were cleaned by sequential sonication in detergent (Decon-90) solution, water, and then propan-2-ol before being stored in water until use. Immediately before use, the coverslip was dried under nitrogen and cleaned with oxygen plasma (Diener Electronic, Femto) for 3 min. A well was created on each coverslip using vacuum grease (Dow Corning high-vacuum grease). A 50 μL amount of SUV stock was diluted 1:1 in buffer (phosphate-buffered saline (PBS) pH 7.4) and added to the chamber immediately. The vesicles were incubated for 60 min before the membranes were washed with degassed Milli-Q, followed by PBS buffer and finally replaced with 1× TE + 14 mM MgCl2 buffer.
GUV Formation
GUVs were prepared by electroformation.39,40 A 100 μL amount of lipid in chloroform (0.786 mg mL–1 DOPC and 0.1 mol % Atto488-DOPE) was applied to the conducting surfaces of two clean ITO-coated slides (Delta Technologies, Loveland, CO, and Sigma-Aldrich, St. Louis, MO) and dried under vacuum for >30 min. The two slides were assembled as a capacitor, with a PDMS spacer of 1 mM thickness used to create a chamber of 1 mL volume between them. This chamber was filled with ∼52 mM sucrose solution (equiosmolar with 1× TE, 14 mM MgCl2 buffer), and the chamber (PDMS) was fastened with binder clips. Electrodes were attached to the ITO slides via adhesive copper tape, and an alternating square wave potential of 1.4 V rms amplitude and 10 Hz frequency was applied for 90 min. The resultant GUVs were transferred using a syringe with large-bore, 21-gauge needle to avoid shearing.
Acknowledgments
This research was supported by FP7Marie Curie Initial Training Network ESCODNA (PID 317110), by Engineering and Physical Sciences Research Council Studentship 1380773 “Dynamic Membrane Structures”, and by European Research Council Starting Grant CoSMiC (PID 309651). A.J.T. was supported by a Royal Society Wolfson Research Merit Award. M.W. and V.R. were supported by the European Research Council (ERC-170 2012-StG-106913, CoSMiC).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.8b07734.
Information on DNA origami design; protocol and plan of apparatus used for preparation of lipid monolayer samples for TEM; supporting data, including confocal z-stacks of unpolymerized and polymerized triskelia on GUVs (PDF)
Epifluorescence movie showing the diffusion of polymerized triskelia on GUVs (AVI)
Epifluorescence movie (AVI)
Epifluorescence movie (AVI)
Author Contributions
C.M.A.J. designed and characterized the DNA origami triskelion nanostructures, including TEM imaging. C.M.A.J. and V.R. studied their interactions with membranes using optical microscopy. M.W. and A.J.T. helped with the design and interpretation of experiments. C.M.A.J. wrote the paper with the assistance of all authors; all authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Zimmerberg J.; Kozlov M. M. How Proteins Produce Cellular Membrane Curvature. Nat. Rev. Mol. Cell Biol. 2006, 7, 9–19. 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- Sleytr U. B.; Schuster B.; Egelseer E.-M.; Pum D. S-Layers: Principles and Applications. FEMS Microbiol. Rev. 2014, 38, 823–864. 10.1111/1574-6976.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouter A.; Gounou C.; Bérat R.; Tan S.; Gallois B.; Granier T.; d’Estaintot B. L.; Pöschl E.; Brachvogel B.; Brisson A. R. Annexin-A5 Assembled into Two-Dimensional Arrays Promotes Cell Membrane Repair. Nat. Commun. 2011, 2, 270. 10.1038/ncomms1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty G. J.; McMahon H. T. Mechanisms of Endocytosis. Annu. Rev. Biochem. 2009, 78, 857–902. 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Jahn R.; Scheller R. H. SNAREs — Engines for Membrane Fusion. Nat. Rev. Mol. Cell Biol. 2006, 7, 631–643. 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Mettlen M.; Pucadyil T.; Ramachandran R.; Schmid S. L. Dissecting Dynamin’s Role in Clathrin-Mediated Endocytosis. Biochem. Soc. Trans. 2009, 37, 1022–1026. 10.1042/BST0371022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocabey S.; Kempter S.; List J.; Xing Y.; Bae W.; Schiffels D.; Shih W. M.; Simmel F. C.; Liedl T. Membrane-Assisted Growth of DNA Origami Nanostructure Arrays. ACS Nano 2015, 9, 3530–3539. 10.1021/acsnano.5b00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czogalla A.; Kauert D. J.; Franquelim H. G.; Uzunova V.; Zhang Y.; Seidel R.; Schwille P. Amphipathic DNA Origami Nanoparticles to Scaffold and Deform Lipid Membrane Vesicles. Angew. Chem., Int. Ed. 2015, 54, 6501–6505. 10.1002/anie.201501173. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Yang Y.; Pincet F.; Llaguno M. C.; Lin C. Placing and Shaping Liposomes with Reconfigurable DNA Nanocages. Nat. Chem. 2017, 9, 653–659. 10.1038/nchem.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkholz O.; Burns J. R.; Richter C. P.; Psathaki O. E.; Howorka S.; Piehler J. Multi-Functional DNA Nanostructures that Puncture and Remodel Lipid Membranes into Hybrid Materials. Nat. Commun. 2018, 9, 1521. 10.1038/s41467-018-02905-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franquelim H. G.; Khmelinskaia A.; Sobczak J.-P.; Dietz H.; Schwille P. Membrane Sculpting by Curved DNA Origami Scaffolds. Nat. Commun. 2018, 9, 811. 10.1038/s41467-018-03198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.; Nathwani B.; Lin C.; Wang J.; Karatekin E.; Pincet F.; Shih W.; Rothman J. E. A Programmable DNA Origami Platform to Organize SNAREs for Membrane Fusion. J. Am. Chem. Soc. 2016, 138, 4439–4447. 10.1021/jacs.5b13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grome M. W.; Zhang Z.; Pincet F.; Lin C. Vesicle Tubulation with Self-Assembling DNA Nanosprings. Angew. Chem., Int. Ed. 2018, 57, 5330–5334. 10.1002/anie.201800141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y.; Endo M.; Sugiyama H. Lipid-Bilayer-Assisted Two-Dimensional Self-Assembly of DNA Origami Nanostructures. Nat. Commun. 2015, 6, 8052. 10.1038/ncomms9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michanek A.; Kristen N.; Höök F.; Nylander T.; Sparr E. RNA and DNA Interactions with Zwitterionic and Charged Lipid Membranes — A DSC and QCM-D Study. Biochim. Biophys. Acta, Biomembr. 2010, 1798, 829–838. 10.1016/j.bbamem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Johnson-Buck A.; Jiang S.; Yan H.; Walter N. G. DNA–Cholesterol Barges as Programmable Membrane-Exploring Agents. ACS Nano 2014, 8, 5641–5649. 10.1021/nn500108k. [DOI] [PubMed] [Google Scholar]
- Khmelinskaia A.; Mucksch J.; Petrov E. P.; Franquelim H. G.; Schwille P. Control of Membrane Binding and Diffusion of Cholesteryl-Modified DNA Origami Nanostructures by DNA Spacers. Langmuir 2018, 34, 14921–14931. 10.1021/acs.langmuir.8b01850. [DOI] [PubMed] [Google Scholar]
- Arnott P. M.; Joshi H.; Aksimentiev A.; Howorka S. Dynamic Interactions Between Lipid-Tethered DNA and Phospholipid Membranes. Langmuir 2018, 34, 15084–15092. 10.1021/acs.langmuir.8b02271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börjesson K.; Lundberg E. P.; Woller J. G.; Nordén B.; Albinsson B. Soft-Surface DNA Nanotechnology: DNA Constructs Anchored and Aligned to Lipid Membrane. Angew. Chem., Int. Ed. 2011, 50, 8312–8315. 10.1002/anie.201103338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avakyan N.; Conway J. W.; Sleiman H. F. Long-Range Ordering of Blunt-Ended DNA Tiles on Supported Lipid Bilayers. J. Am. Chem. Soc. 2017, 139, 12027–12034. 10.1021/jacs.7b06572. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Dietz H.; Liedl T.; Högberg B.; Graf F.; Shih W. M. Self-Assembly of DNA into Nanoscale Three-Dimensional Shapes. Nature 2009, 459, 414–418. 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova N. V.; Deyev I. E.; Petrenko A. G. Clathrin-Mediated Endocytosis and Adaptor Proteins. Acta Naturae 2013, 5, 62–73. 10.32607/20758251-2013-5-3-62-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser J. E.; Keen J. H.; Amende L. M.; Lippoldt R. E.; Prasad K. Deep-Etch Visualization of 27S Clathrin: a Tetrahedral Tetramer. J. Cell Biol. 1987, 105, 1999–2009. 10.1083/jcb.105.5.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeham D. E.; Chen C.-Y.; Greene B.; Hwang P. K.; Brodsky F. M. Clathrin Self-Assembly Involves Coordinated Weak Interactions Favorable for Cellular Regulation. EMBO J. 2003, 22, 4980–4990. 10.1093/emboj/cdg511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins M. K.; McMahon H. T. Snap-Shots of Clathrin-Mediated Endocytosis. Trends Biochem. Sci. 2002, 27, 257–263. 10.1016/S0968-0004(02)02089-3. [DOI] [PubMed] [Google Scholar]
- Ford M. G. J.; Mills I. G.; Peter B. J.; Vallis Y.; Praefcke G. J. K.; Evans P. R.; McMahon H. T. Curvature of Clathrin-Coated Pits Driven by Epsin. Nature 2002, 419, 361–366. 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Ford M. G. J.; Pearse B. M. F.; Higgins M. K.; Vallis Y.; Owen D. J.; Gibson A.; Hopkins C. R.; Evans P. R.; McMahon H. T. Simultaneous Binding of PtdIns(4,5)P2; and Clathrin by AP180 in the Nucleation of Clathrin Lattices on Membranes. Science 2001, 291, 1051–1055. 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- Asadi J.; Ferguson S.; Raja H.; Hacker C.; Marius P.; Ward R.; Pliotas C.; Naismith J.; Lucocq J. Enhanced Imaging of Lipid Rich Nanoparticles Embedded in Methylcellulose Films for Transmission Electron Microscopy Using Mixtures of Heavy Metals. Micron 2017, 99, 40–48. 10.1016/j.micron.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannhauser P. N.; Ungewickell E. J. Reconstitution of Clathrin-Coated Bud and Vesicle Formation with Minimal Components. Nat. Cell Biol. 2012, 14, 634–639. 10.1038/ncb2478. [DOI] [PubMed] [Google Scholar]
- Stachowiak J. C.; Schmid E. M.; Ryan C. J.; Ann H. S.; Sasaki D. Y.; Sherman M. B.; Geissler P. L.; Fletcher D. A.; Hayden C. C. Membrane Bending by Protein-Protein Crowding. Nat. Cell Biol. 2012, 14, 944–949. 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- Snead W. T.; Hayden C. C.; Gadok A. K.; Zhao C.; Lafer E. M.; Rangamani P.; Stachowiak J. C. Membrane Fission by Protein Crowding. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E3258–E3267. 10.1073/pnas.1616199114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B. Mechanisms of Membrane Curvature Sensing. Annu. Rev. Biochem. 2011, 80, 101–123. 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- Bhatia V. K.; Hatzakis N. S.; Stamou D. A Unifying Mechanism Accounts for Sensing of Membrane Curvature by BAR Domains, Amphipathic Helices and Membrane-Anchored Proteins. Semin. Cell Dev. Biol. 2010, 21, 381–390. 10.1016/j.semcdb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Bending Membranes. Nat. Cell Biol. 2012, 14, 906–908. 10.1038/ncb2570. [DOI] [PubMed] [Google Scholar]
- Bell G. I. Models for Specific Adhesion of Cells to Cells. Science 1978, 200, 618–627. 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- McCloskey M. A.; Poo M. Contact-Induced Redistribution of Specific Membrane-Components - Local Accumultaion and Development of Adhesion. J. Cell Biol. 1986, 102, 2185–2196. 10.1083/jcb.102.6.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langecker M.; Arnaut V.; List J.; Simmel F. C. DNA Nanostructures Interacting with Lipid Bilayer Membranes. Acc. Chem. Res. 2014, 47, 1807–1815. 10.1021/ar500051r. [DOI] [PubMed] [Google Scholar]
- Booth D. S.; Avila-Sakar A.; Cheng Y. Visualizing Proteins and Macromolecular Complexes by Negative Stain EM: from Grid Preparation to Image Acquisition. J. Visualized Exp. 2011, 3227. 10.3791/3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelova M. I.; Dimitrov D. S. Liposome Electroformation. Faraday Discuss. Chem. Soc. 1986, 81, 303–311. 10.1039/dc9868100303. [DOI] [Google Scholar]
- Dimova R. Recent Developments in the Field of Bending Rigidity Measurements on Membranes. Adv. Colloid Interface Sci. 2014, 208, 225–234. 10.1016/j.cis.2014.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.