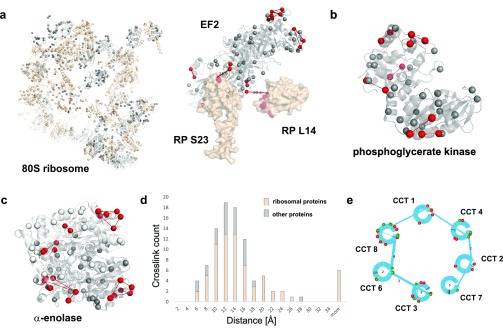
Figure 3.
Application of PhoX to investigate human cell lysates. Cross-linked lysines are depicted as red spheres; non-cross-linked lysines are depicted as gray spheres. (a) Ribosome cryo-EM map (PDB, 4v6x). Proteins found cross-linked are shown in light brown; proteins without cross-links are shown in gray (RNA is not shown). Intra-cross-links mapped on the elongation factor 2 (part of PDB, 4v6x). Inter-cross-links of elongation factor to ribosomal proteins mapped on the ribosome (PDB, 6v6x). (b) Intra-cross-links of phosphoglycerate kinase mapped on its crystal structure (PDB, 3c39) (c) Intra-cross-links of α-enolase mapped on the dimeric crystal structure of human enolase 1 (PDB, 3b97). (d) Histogram of observed Lys–Lys distances in PhoX cross-links on the 80S ribosome (colored in light brown) and other proteins (nucleolin, PDB, 2kkr; nucleophosmin, PDB, 2llh; elongation factor 2, PDB, 4v6x; stress-induced-phosphoprotein 1, PDB, 1ewl; α-enolase, PDB, 3b97; phosphoglycerate kinase, PDB, 3c39). (e) Interaction network found for the TRiC/CCT complex. Proteins of the TRiC/CCT complex are shown as blue circles. Interlinks between the proteins are shown as blue lines; the thickness of the lines indicates the amount of interlinks identified. Green lines in the circles are the lysine positions; red balls indicate phosphorylation sites as extracted from uniprot, and green balls indicate lysines involved in interlinks.