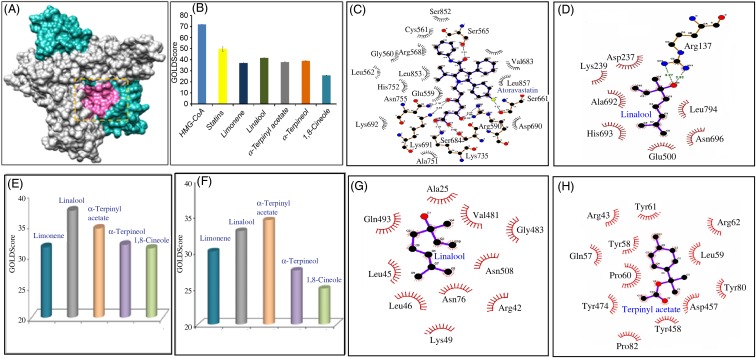
Fig. 10.
(A) Three-dimensional surface view of one of the dimer structures of 3-hydroxy-3-methyl-glutaryl-CoA (HMG-CoA) receptor (Protein Data Bank ID: 1HWK) where two chains are marked in grey and cyan and the substrate/inhibitor binding pocket is shown in pink. (B) Average molecular docking score represented as GOLDScore is plotted for each active ingredient of the cardamom extract when docked at the substrate/inhibitor binding site. GOLD derived rescores of the substrate (HMG-CoA) and inhibitors (statins) bound to HMG-CoA receptor protein are also plotted for comparison purposes. Values are means, with standard deviations represented by vertical bars. (C, D) LigPlot representations of the interacting residues and the probable interaction types for one of the known inhibitor (atorvastatin) and the highest-scoring active ingredient (linalool), respectively. (E, F) Average molecular docking scores represented as GOLDScore are plotted for each active ingredient of the cardamom extract when docked at the predicted binding sites, consensus pocket 2 and 3, respectively. (G, H) LigPlot representations of the interacting residues and the probable interaction types derived from linalool and α-terpinyl acetate docked at the predicted binding sites, consensus pocket 2 and 3, respectively.