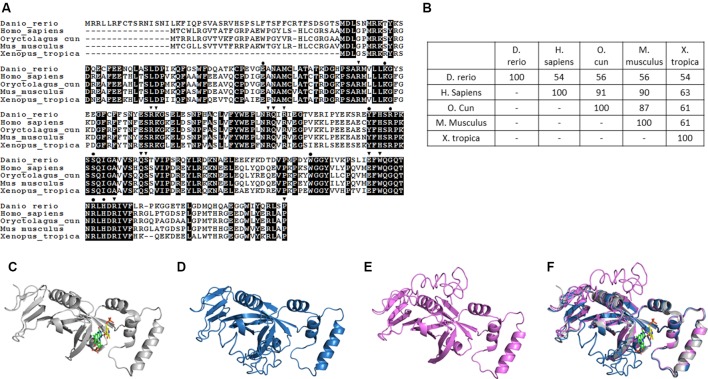
Figure 2.
Structural and phylogenic comparison of zebrafish Pnpo with enzymes from four different sources. The amino acid sequences of PNPOs from the indicated species were analyzed and compared for functional domains and evolutionary conservation. (A) The peptide sequences of PNPOs from the indicated species were aligned. The shaded letters indicate the amino acids that are identical among compared species. Dots indicate the PNP/PLP binding sites, and arrowheads indicate the FMN binding sites. (B) A phylogenetic table shows the identity between compared enzymes, revealing the evolutionary relationships of PNPOs among the compared species. (C) The crystal structure of experimental hPNPO (Musayev et al., 2003), (D) the simulated structures of hPNPO, and (E) zPnpo are shown in ribbon diagram. (F) Superposed experimental hPNPO crystal structure (grey) and simulated zPnpo (magenta) and hPNPO (cyan) structures. The simulated structures are obtained using I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/). The compared peptide sequences include zebrafish Pnpo (NP_001243107.1); human PNPO (NP_060599.1); rabbit PNPO (XP_002719371.1); mouse PNPO (NP_598782.1); and Xenopus Pnpo (NP_001120016.1). PNPO, pyridoxine 5′-phosphate oxidase; PLP, pyridoxal 5′-phosphate.