Figure 1. Small Protein Discovery and Characterization Pipeline Applied to HMPI-II Metagenomic Data.
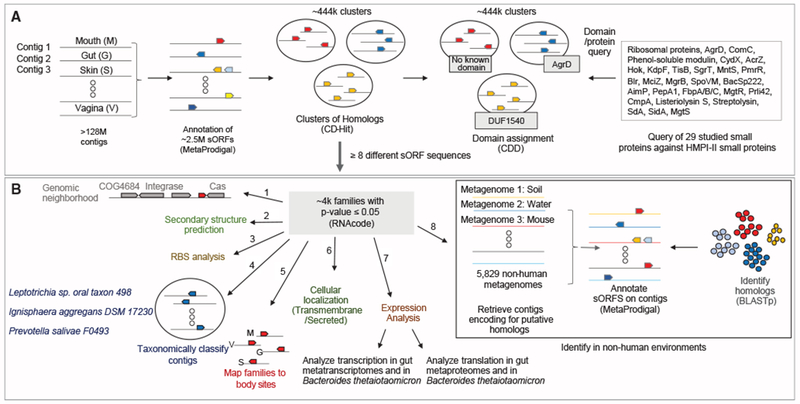
(A) Identification of 29 known small proteins in HMPI-II metagenomes. More than 128 million contigs were annotated using MetaProdigal with a lower size limit of five amino acids. The small proteins were then clustered using CD-Hit based on amino acid similarity and protein length. Representatives of each of the ~444,000 clusters were queried against the Conserved Domain Database (CDD), to assign domains to clusters. The list of CDD domains was then queried for the small known proteins that have an assigned domain. Known small proteins that do not have an assigned domain or that failed the domain search were queried against HMPI-II small proteins using BLASTp.
(B) Identification and characterization of HMPI-II small proteins. RNAcode was used to assign p values to the ~444,000 clusters. The following analyses were conducted on the ~4,000 protein families whose p value was ≤0.05. (1) Identification of neighboring genes on longest contig associated with each family. (2) Prediction of secondary structure. (3) Analysis of ribosomal binding sites (RBS) upstream of the small genes. (4) Taxonomic classification of contigs encoding each of the small protein families. (5) Assignment of small protein families to body sites. M-mouth; V - vagina; G - gut; S - skin. (6) Prediction of signal peptide and transmembrane domains to assign likely cellular localization. (7) Analysis of expression of the small genes using metatranscriptomic, metaproteomic datasets as well as Bacteroides thetaiotaomicron transcriptomics and proteomics. (8) Identification of homologs of small protein families in non-human metagenomes.
See also Figures S1, S2, and S7, Tables S1, S2, S3, and S4, and Data S1 and S2.
