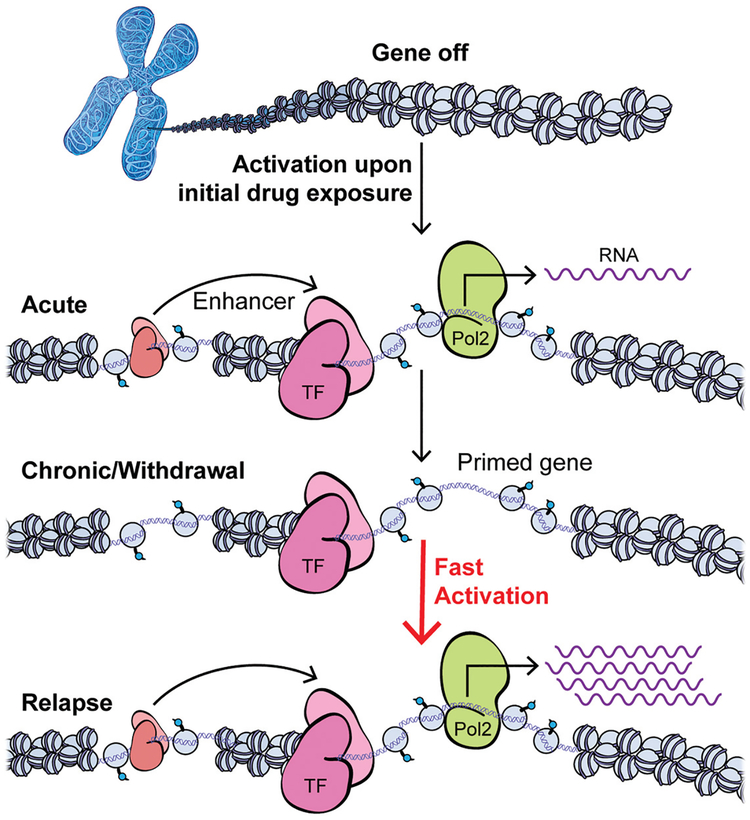
Figure 3.
Time-dependent control of gene regulation by drug exposure via epigenetic mechanisms. Chromatin mechanisms not only regulate the acute transcriptional response to drug exposure but further mediate latent effects at many genes that alter their inducibility to future stimuli. Transcription initiates within core promoters at the transcriptional start sites (TSSs) at 5′ ends of genes, which recruit RNA polymerase II (Pol2) and determine the accurate initiation position and direction of transcription. Efficient transcription is supported by enhancer elements (located at some distance from their target genes) that contain transcription factor (TF; “salmon”) binding sites and recruit a combination of TFs with a variety of cofactors to exert their overall regulatory function to control transcription from the targeted core promoter. Neuronal activation upon the initial drug exposure triggers intracellular signaling cascades that activate TFs and many other nuclear targets, including chromatin-regulatory proteins that modify histones and other proteins to regulate DNA accessibility, as well as transcription initiation and elongation. Early evidence suggests that chronic drug exposure causes extremely stable changes at the chromatin level that underlie transcriptional priming (shown) and desensitization (not shown) linked to drug addiction. Such gene priming/desensitization may remain latent during periods of withdrawal, when certain TFs and cofactors are bound to accessible chromatin without changing the steady state mRNA levels. However, future context and/or drug re-exposure can up-regulate a primed gene much faster and to a greater extent, based on the epigenetic changes induced by previous chronic drug exposure at regulatory promoter and enhancer regions.